Suppressors of transcriptional transgenic silencing in Chlamydomonas are sensitive to DNA-damaging agents and reactivate transposable elements
- PMID: 11782532
- PMCID: PMC117432
- DOI: 10.1073/pnas.022392999
Suppressors of transcriptional transgenic silencing in Chlamydomonas are sensitive to DNA-damaging agents and reactivate transposable elements
Abstract
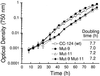
In the unicellular green alga Chlamydomonas reinhardtii, the epigenetic silencing of transgenes occurs, as in land plants, at both the transcriptional and posttranscriptional levels. In the case of single-copy transgenes, transcriptional silencing takes place without detectable cytosine methylation of the introduced DNA. We have isolated two mutant strains, Mut-9 and Mut-11, that reactivate expression of a transcriptionally silenced single-copy transgene. These suppressors are deficient in the repression of a DNA transposon and a retrotransposon-like element. In addition, the mutants show enhanced sensitivity to DNA-damaging agents, particularly radiomimetic chemicals inducing DNA double-strand breaks. All of these phenotypes are much more prominent in a double mutant strain. These observations suggest that multiple partly redundant epigenetic mechanisms are involved in the repression of transgenes and transposons in eukaryotes, presumably as components of a system that evolved to preserve genomic stability. Our results also raise the possibility of mechanistic connections between epigenetic transcriptional silencing and DNA double-strand break repair.
Figures
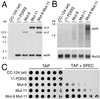
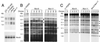
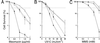
 , wild-type CC-124; ●, Mut-9; Δ, Mut-11;
♦, Mut-9 Mut-11. (A) Survival of the mutants
and wild-type C. reinhardtii grown on TAP medium
containing increasing concentrations of bleomycin. (B)
Survival of the mutants and wild-type C. reinhardtii
exposed to increasing doses of UV-C irradiation under
nonphotoreactivating conditions. (C) Survival of the
mutants and wild-type C. reinhardtii grown on TAP medium
containing increasing concentrations of methyl methanesulfonate
(MMS).
, wild-type CC-124; ●, Mut-9; Δ, Mut-11;
♦, Mut-9 Mut-11. (A) Survival of the mutants
and wild-type C. reinhardtii grown on TAP medium
containing increasing concentrations of bleomycin. (B)
Survival of the mutants and wild-type C. reinhardtii
exposed to increasing doses of UV-C irradiation under
nonphotoreactivating conditions. (C) Survival of the
mutants and wild-type C. reinhardtii grown on TAP medium
containing increasing concentrations of methyl methanesulfonate
(MMS).Similar articles
-
Functional specialization of Chlamydomonas reinhardtii cytosolic thioredoxin h1 in the response to alkylation-induced DNA damage.Eukaryot Cell. 2005 Feb;4(2):262-73. doi: 10.1128/EC.4.2.262-273.2005. Eukaryot Cell. 2005. PMID: 15701788 Free PMC article.
-
A WD40-repeat containing protein, similar to a fungal co-repressor, is required for transcriptional gene silencing in Chlamydomonas.Plant J. 2002 Jul;31(1):25-36. doi: 10.1046/j.1365-313x.2002.01331.x. Plant J. 2002. PMID: 12100480
-
Unstable RNAi effects through epigenetic silencing of an inverted repeat transgene in Chlamydomonas reinhardtii.Genetics. 2008 Dec;180(4):1927-44. doi: 10.1534/genetics.108.092395. Epub 2008 Oct 1. Genetics. 2008. PMID: 18832355 Free PMC article.
-
Gene silencing: double-stranded RNA mediated mRNA degradation and gene inactivation.Cell Res. 2001 Sep;11(3):181-6. doi: 10.1038/sj.cr.7290084. Cell Res. 2001. PMID: 11642402 Review.
-
Differentiation of epigenetic modifications between transposons and genes.Curr Opin Plant Biol. 2011 Feb;14(1):81-7. doi: 10.1016/j.pbi.2010.08.017. Epub 2010 Sep 23. Curr Opin Plant Biol. 2011. PMID: 20869294 Review.
Cited by
-
Good News for Nuclear Transgene Expression in Chlamydomonas.Cells. 2019 Nov 28;8(12):1534. doi: 10.3390/cells8121534. Cells. 2019. PMID: 31795196 Free PMC article. Review.
-
Functional specialization of Chlamydomonas reinhardtii cytosolic thioredoxin h1 in the response to alkylation-induced DNA damage.Eukaryot Cell. 2005 Feb;4(2):262-73. doi: 10.1128/EC.4.2.262-273.2005. Eukaryot Cell. 2005. PMID: 15701788 Free PMC article.
-
Osmotic stress induces phosphorylation of histone H3 at threonine 3 in pericentromeric regions of Arabidopsis thaliana.Proc Natl Acad Sci U S A. 2015 Jul 7;112(27):8487-92. doi: 10.1073/pnas.1423325112. Epub 2015 Jun 22. Proc Natl Acad Sci U S A. 2015. PMID: 26100864 Free PMC article.
-
Increasing lipid production in Chlamydomonas reinhardtii through genetic introduction for the overexpression of glyceraldehyde-3-phosphate dehydrogenase.Front Bioeng Biotechnol. 2024 Apr 19;12:1396127. doi: 10.3389/fbioe.2024.1396127. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 38707501 Free PMC article.
-
Mu killer causes the heritable inactivation of the Mutator family of transposable elements in Zea mays.Genetics. 2003 Oct;165(2):781-97. doi: 10.1093/genetics/165.2.781. Genetics. 2003. PMID: 14573488 Free PMC article.
References
-
- Wolffe A P, Matzke M A. Science. 1999;286:481–486. - PubMed
-
- Finnegan E J, Peacock W J, Dennis E S. Curr Opin Genet Dev. 2000;10:217–223. - PubMed
-
- Habu Y, Kakutani T, Paszkowski J. Curr Opin Genet Dev. 2001;11:215–220. - PubMed
-
- Matzke M, Matzke A J M, Kooter J M. Science. 2001;293:1080–1083. - PubMed
-
- Vaucheret H, Fagard M. Trends Genet. 2001;17:29–35. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

