The chloroplastic GrpE homolog of Chlamydomonas: two isoforms generated by differential splicing
- PMID: 11752390
- PMCID: PMC139491
- DOI: 10.1105/tpc.010202
The chloroplastic GrpE homolog of Chlamydomonas: two isoforms generated by differential splicing
Abstract
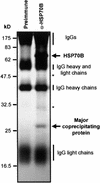
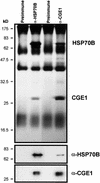
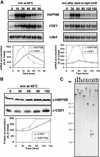
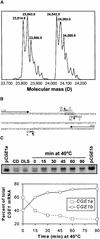
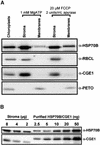
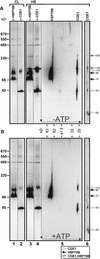
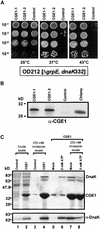
In eubacteria and mitochondria, Hsp70 chaperone activity is controlled by the nucleotide exchange factor GrpE. We have identified the chloroplastic GrpE homolog of Chlamydomonas, CGE1, as an approximately 26-kD protein coimmunoprecipitating with the stromal HSP70B protein. When expressed in Escherichia coli, CGE1 can functionally replace GrpE and interacts physically with DnaK. CGE1 is encoded by a single-copy gene that is induced strongly by heat shock and slightly by light. Alternative splicing generates two isoforms that differ only by two residues in the N-terminal part. The larger form is synthesized preferentially during heat shock, whereas the smaller one dominates at lower temperatures. Fractions of both HSP70B and CGE1 associate with chloroplast membranes in an ATP-sensitive manner. By colorless native PAGE and pulse labeling, CGE1 monomers were found to assemble rapidly into dimers and tetramers. In addition, CGE1 was found to form ATP-sensitive complexes with HSP70B of approximately 230 and approximately 120 kD, the latter increasing dramatically after heat shock.
Figures



Similar articles
-
The chloroplast DnaJ homolog CDJ1 of Chlamydomonas reinhardtii is part of a multichaperone complex containing HSP70B, CGE1, and HSP90C.Plant Physiol. 2008 Dec;148(4):2070-82. doi: 10.1104/pp.108.127944. Epub 2008 Oct 17. Plant Physiol. 2008. PMID: 18931144 Free PMC article.
-
The NH2-terminal domain of the chloroplast GrpE homolog CGE1 is required for dimerization and cochaperone function in vivo.J Biol Chem. 2007 Apr 13;282(15):11317-28. doi: 10.1074/jbc.M608854200. Epub 2007 Feb 8. J Biol Chem. 2007. PMID: 17289679
-
Assistance for a chaperone: Chlamydomonas HEP2 activates plastidic HSP70B for cochaperone binding.J Biol Chem. 2008 Jun 13;283(24):16363-73. doi: 10.1074/jbc.M708431200. Epub 2008 Apr 17. J Biol Chem. 2008. PMID: 18420590
-
The chloroplast HSP70B-CDJ2-CGE1 chaperones catalyse assembly and disassembly of VIPP1 oligomers in Chlamydomonas.Plant J. 2007 Apr;50(2):265-77. doi: 10.1111/j.1365-313X.2007.03047.x. Epub 2007 Mar 12. Plant J. 2007. PMID: 17355436
-
The Escherichia coli chaperones involved in DNA replication.Philos Trans R Soc Lond B Biol Sci. 1993 Mar 29;339(1289):271-7; discussion 277-8. doi: 10.1098/rstb.1993.0025. Philos Trans R Soc Lond B Biol Sci. 1993. PMID: 8098531 Review.
Cited by
-
Regulation by Light of Chemotaxis to Nitrite during the Sexual Life Cycle in Chlamydomonas reinhardtii.Plants (Basel). 2014 Feb 26;3(1):113-27. doi: 10.3390/plants3010113. Plants (Basel). 2014. PMID: 27135494 Free PMC article.
-
Defects in the cytochrome b6/f complex prevent light-induced expression of nuclear genes involved in chlorophyll biosynthesis.Plant Physiol. 2006 Jul;141(3):1128-37. doi: 10.1104/pp.106.081059. Epub 2006 May 5. Plant Physiol. 2006. PMID: 16679422 Free PMC article.
-
Two Arabidopsis Chloroplast GrpE Homologues Exhibit Distinct Biological Activities and Can Form Homo- and Hetero-Oligomers.Front Plant Sci. 2020 Jan 22;10:1719. doi: 10.3389/fpls.2019.01719. eCollection 2019. Front Plant Sci. 2020. PMID: 32038688 Free PMC article.
-
Complexome profiling on the Chlamydomonas lpa2 mutant reveals insights into PSII biogenesis and new PSII associated proteins.J Exp Bot. 2022 Jan 5;73(1):245-262. doi: 10.1093/jxb/erab390. J Exp Bot. 2022. PMID: 34436580 Free PMC article.
-
VIPP1 rods engulf membranes containing phosphatidylinositol phosphates.Sci Rep. 2019 Jun 19;9(1):8725. doi: 10.1038/s41598-019-44259-3. Sci Rep. 2019. PMID: 31217458 Free PMC article.
References
-
- Asamizu, E., Nakamura, Y., Sato, S., Fukuzawa, H., and Tabata, S. (1999). A large scale structural analysis of cDNAs in a unicellular green alga, Chlamydomonas reinhardtii: Generation of 3433 non-redundant expressed sequence tags. DNA Res. 6, 369–373. - PubMed
-
- Azem, A., Oppliger, W., Lustig, A., Jenö, P., Feifel, B., Schatz, G., and Horst, M. (1997). The mitochondrial hsp70 chaperone system: Effect of adenine nucleotides, peptide substrate, and mGrpE on the oligomeric state of mhsp70. J. Biol. Chem. 272, 20901–20906. - PubMed
-
- Ballinger, C.A., Connell, P., Wu, Y., Hu, Z., Thompson, L.J., Yin, L.-Y., and Patterson, C. (1999). Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol. Cell. Biol. 19, 4535–4545. - PMC - PubMed
-
- Beckmann, R.P., Mizzen, L.A., and Welch, W.J. (1990). Interaction of Hsp 70 with newly synthesized proteins: Implications for protein folding and assembly. Science 248, 850–854. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources

