MDB: the Metalloprotein Database and Browser at The Scripps Research Institute
- PMID: 11752342
- PMCID: PMC99158
- DOI: 10.1093/nar/30.1.379
MDB: the Metalloprotein Database and Browser at The Scripps Research Institute
Abstract
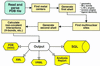
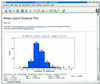
The Metalloprotein Database and Browser (MDB; http://metallo.scripps.edu) at The Scripps Research Institute is a web-accessible resource for metalloprotein research. It offers the scientific community quantitative information on geometrical parameters of metal-binding sites in protein structures available from the Protein Data Bank (PDB). The MDB also offers analytical tools for the examination of trends or patterns in the indexed metal-binding sites. A user can perform interactive searches, metal-site structure visualization (via a Java applet), and analysis of the quantitative data by accessing the MDB through a web browser without requiring an external application or platform-dependent plugin. The MDB also has a non-interactive interface with which other web sites and network-aware applications can seamlessly incorporate data or statistical analysis results from metal-binding sites. The information contained in the MDB is periodically updated with automated algorithms that find and index metal sites from new protein structures released by the PDB.
Figures
Similar articles
-
Development of tools and database for analysis of metal binding sites in protein.Protein Pept Lett. 2010 Jun;17(6):765-73. doi: 10.2174/092986610791190246. Protein Pept Lett. 2010. PMID: 20205657
-
MESPEUS: a database of metal coordination groups in proteins.Nucleic Acids Res. 2024 Jan 5;52(D1):D483-D493. doi: 10.1093/nar/gkad1009. Nucleic Acids Res. 2024. PMID: 37941148 Free PMC article.
-
InterMetalDB: A Database and Browser of Intermolecular Metal Binding Sites in Macromolecules with Structural Information.J Proteome Res. 2021 Apr 2;20(4):1889-1901. doi: 10.1021/acs.jproteome.0c00906. Epub 2021 Jan 27. J Proteome Res. 2021. PMID: 33502860 Free PMC article.
-
Protein structure databases with new web services for structural biology and biomedical research.Brief Bioinform. 2008 Jul;9(4):276-85. doi: 10.1093/bib/bbn015. Epub 2008 Apr 22. Brief Bioinform. 2008. PMID: 18430752 Review.
-
Metalloprotein design.Curr Opin Biotechnol. 1996 Aug;7(4):437-41. doi: 10.1016/s0958-1669(96)80121-2. Curr Opin Biotechnol. 1996. PMID: 8768904 Review.
Cited by
-
A less-biased analysis of metalloproteins reveals novel zinc coordination geometries.Proteins. 2015 Aug;83(8):1470-87. doi: 10.1002/prot.24834. Epub 2015 Jun 13. Proteins. 2015. PMID: 26009987 Free PMC article.
-
Conserved structural chemistry for incision activity in structurally non-homologous apurinic/apyrimidinic endonuclease APE1 and endonuclease IV DNA repair enzymes.J Biol Chem. 2013 Mar 22;288(12):8445-8455. doi: 10.1074/jbc.M112.422774. Epub 2013 Jan 25. J Biol Chem. 2013. PMID: 23355472 Free PMC article.
-
Structural characterization of the zinc binding domain in cytosolic PSD-95 interactor (cypin): Role of zinc binding in guanine deamination and dendrite branching.Proteins. 2008 Feb 15;70(3):873-81. doi: 10.1002/prot.21683. Proteins. 2008. PMID: 17803218 Free PMC article.
-
The protective role of ascorbic acid in the hepatotoxicity of cadmium and mercury in rabbits.Environ Sci Pollut Res Int. 2019 May;26(14):14087-14096. doi: 10.1007/s11356-019-04620-5. Epub 2019 Mar 9. Environ Sci Pollut Res Int. 2019. PMID: 30852747
-
Evidence for a polynuclear metal ion binding site in the catalytic domain of ribonuclease P RNA.EMBO J. 2002 May 1;21(9):2253-62. doi: 10.1093/emboj/21.9.2253. EMBO J. 2002. PMID: 11980722 Free PMC article.
References
-
- Hooft R.W.W., Sander,C. and Vriend,G. (1996) Verification of protein structures: side-chain planarity. J. Appl. Cryst., 29, 714–716.