Solution structure of conserved AGNN tetraloops: insights into Rnt1p RNA processing
- PMID: 11743001
- PMCID: PMC125334
- DOI: 10.1093/emboj/20.24.7250
Solution structure of conserved AGNN tetraloops: insights into Rnt1p RNA processing
Abstract
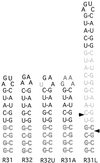
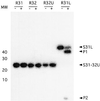
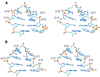
Rnt1p, the yeast orthologue of RNase III, cleaves rRNAs, snRNAs and snoRNAs at a stem capped with conserved AGNN tetraloop. Here we show that 9 bp long stems ending with AGAA or AGUC tetraloops bind to Rnt1p and direct specific but sequence-independent RNA cleavage when provided with stems longer than 13 bp. The solution structures of these two tetraloops reveal a common fold for the terminal loop stabilized by non-canonical A-A or A-C pairs and extensive base stacking. The conserved nucleotides are stacked at the 5' side of the loop, exposing their Watson-Crick and Hoogsteen faces for recognition by Rnt1p. These results indicate that yeast RNase III recognizes the fold of a conserved single-stranded tetraloop to direct specific dsRNA cleavage.
Figures


Similar articles
-
Recognition modes of RNA tetraloops and tetraloop-like motifs by RNA-binding proteins.Wiley Interdiscip Rev RNA. 2014 Jan-Feb;5(1):49-67. doi: 10.1002/wrna.1196. Epub 2013 Oct 3. Wiley Interdiscip Rev RNA. 2014. PMID: 24124096 Free PMC article. Review.
-
Structure of an AAGU tetraloop and its contribution to substrate selection by yeast RNase III.J Mol Biol. 2006 Oct 20;363(2):322-31. doi: 10.1016/j.jmb.2006.08.029. Epub 2006 Aug 16. J Mol Biol. 2006. PMID: 16979185
-
A novel family of RNA tetraloop structure forms the recognition site for Saccharomyces cerevisiae RNase III.EMBO J. 2001 Dec 17;20(24):7240-9. doi: 10.1093/emboj/20.24.7240. EMBO J. 2001. PMID: 11743000 Free PMC article.
-
Recognition of a conserved class of RNA tetraloops by Saccharomyces cerevisiae RNase III.Proc Natl Acad Sci U S A. 2000 Mar 28;97(7):3142-7. doi: 10.1073/pnas.97.7.3142. Proc Natl Acad Sci U S A. 2000. PMID: 10716739 Free PMC article.
-
Structure and function of Rnt1p: An alternative to RNAi for targeted RNA degradation.Wiley Interdiscip Rev RNA. 2019 May;10(3):e1521. doi: 10.1002/wrna.1521. Epub 2018 Dec 11. Wiley Interdiscip Rev RNA. 2019. PMID: 30548404 Review.
Cited by
-
Conformational adaptation of UNCG loops upon crowding.RNA. 2019 Nov;25(11):1522-1531. doi: 10.1261/rna.072694.119. Epub 2019 Aug 19. RNA. 2019. PMID: 31427457 Free PMC article.
-
Structural basis for recognition of the AGNN tetraloop RNA fold by the double-stranded RNA-binding domain of Rnt1p RNase III.Proc Natl Acad Sci U S A. 2004 Jun 1;101(22):8307-12. doi: 10.1073/pnas.0402627101. Epub 2004 May 18. Proc Natl Acad Sci U S A. 2004. PMID: 15150409 Free PMC article.
-
Dynamics of strand slippage in DNA hairpins formed by CAG repeats: roles of sequence parity and trinucleotide interrupts.Nucleic Acids Res. 2020 Mar 18;48(5):2232-2245. doi: 10.1093/nar/gkaa036. Nucleic Acids Res. 2020. PMID: 31974547 Free PMC article.
-
Recognition modes of RNA tetraloops and tetraloop-like motifs by RNA-binding proteins.Wiley Interdiscip Rev RNA. 2014 Jan-Feb;5(1):49-67. doi: 10.1002/wrna.1196. Epub 2013 Oct 3. Wiley Interdiscip Rev RNA. 2014. PMID: 24124096 Free PMC article. Review.
-
Short RNA guides cleavage by eukaryotic RNase III.PLoS One. 2007 May 30;2(5):e472. doi: 10.1371/journal.pone.0000472. PLoS One. 2007. PMID: 17534422 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

