Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis
- PMID: 11742998
- PMCID: PMC125340
- DOI: 10.1093/emboj/20.24.7220
Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis
Abstract
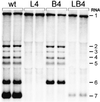
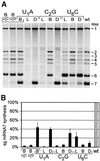
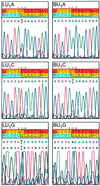
Nidovirus subgenomic mRNAs contain a leader sequence derived from the 5' end of the genome fused to different sequences ('bodies') derived from the 3' end. Their generation involves a unique mechanism of discontinuous subgenomic RNA synthesis that resembles copy-choice RNA recombination. During this process, the nascent RNA strand is transferred from one site in the template to another, during either plus or minus strand synthesis, to yield subgenomic RNA molecules. Central to this process are transcription-regulating sequences (TRSs), which are present at both template sites and ensure the fidelity of strand transfer. Here we present results of a comprehensive co-variation mutagenesis study of equine arteritis virus TRSs, demonstrating that discontinuous RNA synthesis depends not only on base pairing between sense leader TRS and antisense body TRS, but also on the primary sequence of the body TRS. While the leader TRS merely plays a targeting role for strand transfer, the body TRS fulfils multiple functions. The sequences of mRNA leader-body junctions of TRS mutants strongly suggested that the discontinuous step occurs during minus strand synthesis.
Figures


Similar articles
-
The stability of the duplex between sense and antisense transcription-regulating sequences is a crucial factor in arterivirus subgenomic mRNA synthesis.J Virol. 2003 Jan;77(2):1175-83. doi: 10.1128/jvi.77.2.1175-1183.2003. J Virol. 2003. PMID: 12502834 Free PMC article.
-
Regulation of relative abundance of arterivirus subgenomic mRNAs.J Virol. 2004 Aug;78(15):8102-13. doi: 10.1128/JVI.78.15.8102-8113.2004. J Virol. 2004. PMID: 15254182 Free PMC article.
-
Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences.Proc Natl Acad Sci U S A. 1999 Oct 12;96(21):12056-61. doi: 10.1073/pnas.96.21.12056. Proc Natl Acad Sci U S A. 1999. PMID: 10518575 Free PMC article.
-
Nidovirus transcription: how to make sense...?J Gen Virol. 2006 Jun;87(Pt 6):1403-1421. doi: 10.1099/vir.0.81611-0. J Gen Virol. 2006. PMID: 16690906 Review.
-
A new model for coronavirus transcription.Adv Exp Med Biol. 1998;440:215-9. doi: 10.1007/978-1-4615-5331-1_26. Adv Exp Med Biol. 1998. PMID: 9782283 Review.
Cited by
-
PRRSV structure, replication and recombination: Origin of phenotype and genotype diversity.Virology. 2015 May;479-480:475-86. doi: 10.1016/j.virol.2015.02.012. Epub 2015 Mar 7. Virology. 2015. PMID: 25759097 Free PMC article. Review.
-
Molecular characterization of a novel coronavirus associated with epizootic catarrhal enteritis (ECE) in ferrets.Virology. 2006 May 25;349(1):164-74. doi: 10.1016/j.virol.2006.01.031. Epub 2006 Feb 24. Virology. 2006. PMID: 16499943 Free PMC article.
-
Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1''-phosphate dephosphorylation by a conserved domain of nsP3.Structure. 2005 Nov;13(11):1665-75. doi: 10.1016/j.str.2005.07.022. Structure. 2005. PMID: 16271890 Free PMC article.
-
Genome organization of the SARS-CoV.Genomics Proteomics Bioinformatics. 2003 Aug;1(3):226-35. doi: 10.1016/s1672-0229(03)01028-3. Genomics Proteomics Bioinformatics. 2003. PMID: 15629035 Free PMC article.
-
Role of nucleotides immediately flanking the transcription-regulating sequence core in coronavirus subgenomic mRNA synthesis.J Virol. 2005 Feb;79(4):2506-16. doi: 10.1128/JVI.79.4.2506-2516.2005. J Virol. 2005. PMID: 15681451 Free PMC article.
References
-
- Aaziz R. and Tepfer,M. (1999) Recombination in RNA viruses and in virus-resistant transgenic plants. J. Gen. Virol., 80, 1339–1346. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

