Strong natural pausing by RNA polymerase II within 10 bases of transcription start may result in repeated slippage and reextension of the nascent RNA
- PMID: 11739720
- PMCID: PMC134219
- DOI: 10.1128/MCB.22.1.30-40.2002
Strong natural pausing by RNA polymerase II within 10 bases of transcription start may result in repeated slippage and reextension of the nascent RNA
Abstract
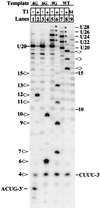
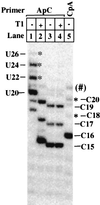
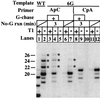
We find that immediately following transcript initiation, RNA polymerase II pauses at several locations even in the presence of relatively high (200 microM) levels of nucleoside triphosphates. Strong pauses with half-lives of >30 s were observed at +7, +18/19, and about +25 on the template used in these experiments. We show that the strong pause at +7, after the synthesis of 5'-ACUCUCU, leads to repeated cycles of upstream slippage of the RNA-DNA hybrid followed by re-pairing with the DNA and continued RNA synthesis. The resulting transcripts are 2, 4, and 6 bases longer than predicted by the template sequence. Slippage is efficient when transcription is primed with the +1/+2 (ApC) dinucleotide, and it occurs at even higher levels with the +2/+3 primer (CpU). Slippage can occur at high levels with ATP initiation, but priming with CpA (-1/+1) supports very little slippage. This latter result is not simply an effect of transcript length at the point of pausing. Slippage can also occur with a second template on which the polymerase can be paused after synthesizing ACUCU. Slippage is not reduced by an ATP analog that blocks promoter escape, but it is inhibited by substitution of 5Br-U for U in the RNA. Our results reveal an unexpected flexibility of RNA polymerase II ternary complexes during the very early stage of transcription, and they suggest that initiation at different locations within the same promoter gives rise to transcription complexes with different properties.
Figures
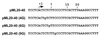



Similar articles
-
Transcription initiation by RNA polymerase II in vitro. At least two nucleotides must be added to form a stable ternary complex.J Biol Chem. 1987 Jan 5;262(1):289-97. J Biol Chem. 1987. PMID: 2432060
-
In vitro studies of transcript initiation by Escherichia coli RNA polymerase. 1. RNA chain initiation, abortive initiation, and promoter escape at three bacteriophage promoters.Biochemistry. 2003 Apr 8;42(13):3777-86. doi: 10.1021/bi026954e. Biochemistry. 2003. PMID: 12667069
-
In vitro studies of transcript initiation by Escherichia coli RNA polymerase. 3. Influences of individual DNA elements within the promoter recognition region on abortive initiation and promoter escape.Biochemistry. 2003 Apr 8;42(13):3798-811. doi: 10.1021/bi026962v. Biochemistry. 2003. PMID: 12667071
-
Promoter escape by RNA polymerase II.Biochim Biophys Acta. 2002 Sep 13;1577(2):208-223. doi: 10.1016/s0167-4781(02)00453-0. Biochim Biophys Acta. 2002. PMID: 12213653 Review.
-
Pause & go: from the discovery of RNA polymerase pausing to its functional implications.Curr Opin Cell Biol. 2017 Jun;46:72-80. doi: 10.1016/j.ceb.2017.03.002. Epub 2017 Mar 28. Curr Opin Cell Biol. 2017. PMID: 28363125 Free PMC article. Review.
Cited by
-
Mediator-regulated transcription through the +1 nucleosome.Mol Cell. 2012 Dec 28;48(6):837-48. doi: 10.1016/j.molcel.2012.10.009. Epub 2012 Nov 15. Mol Cell. 2012. PMID: 23159738 Free PMC article.
-
The initiation-elongation transition: lateral mobility of RNA in RNA polymerase II complexes is greatly reduced at +8/+9 and absent by +23.Proc Natl Acad Sci U S A. 2003 May 13;100(10):5700-5. doi: 10.1073/pnas.1037057100. Epub 2003 Apr 28. Proc Natl Acad Sci U S A. 2003. PMID: 12719526 Free PMC article.
-
RNA polymerase II complexes in the very early phase of transcription are not susceptible to TFIIS-induced exonucleolytic cleavage.Nucleic Acids Res. 2002 Jun 1;30(11):2290-8. doi: 10.1093/nar/30.11.2290. Nucleic Acids Res. 2002. PMID: 12034815 Free PMC article.
-
A novel, non-radioactive eukaryotic in vitro transcription assay for sensitive quantification of RNA polymerase II activity.BMC Mol Biol. 2014 Apr 3;15:7. doi: 10.1186/1471-2199-15-7. BMC Mol Biol. 2014. PMID: 24694320 Free PMC article.
-
Regulation of gene expression by reiterative transcription.Curr Opin Microbiol. 2011 Apr;14(2):142-7. doi: 10.1016/j.mib.2011.01.012. Epub 2011 Feb 19. Curr Opin Microbiol. 2011. PMID: 21334966 Free PMC article. Review.
References
-
- Borukhov, S., V. Sagitov, C. A. Josaitis, R. L. Gourse, and A. Goldfarb. 1993. Two modes of transcription initiation in vitro at the rmB P1 promoter of Escherichia coli. J. Biol. Chem. 268:23477–23482. - PubMed
-
- Cowie, A., P. Jat, and R. Kamen. 1982. Determination of sequences at the capped 5′ ends of polyoma virus early region transcripts synthesized in vivo and in vitro demonstrates an unusual microheterogeneity. J. Mol. Biol. 159:225–255. - PubMed
-
- Dedrick, R. L., C. M. Kane, and M. J. Chamberlin. 1987. Purified RNA polymerase II recognizes specific termination sites during transcription in vitro. J. Biol. Chem. 262:9098–9108. - PubMed
-
- Dvir, A., J. W. Conaway, and R. C. Conaway. 2001. Mechanism of transcription initiation and promoter escape by RNA polymerase II. Curr. Opin. Genet. Dev. 11:209–214. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
