The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions
- PMID: 11739701
- PMCID: PMC135715
- DOI: 10.1128/jvi.76.1.364-378.2002
The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions
Abstract
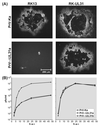
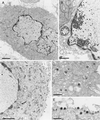
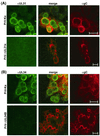
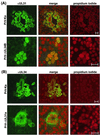
A 2.6-kbp fragment of the pseudorabies virus (PrV) genome was sequenced and shown to contain the homologues of the highly conserved herpesvirus genes UL31 and UL32. By use of a monospecific antiserum, the UL31 gene product was identified as a nuclear protein with an apparent molecular mass of 29 kDa. For functional analysis, UL31 was deleted by mutagenesis in Escherichia coli of an infectious full-length clone of the PrV genome. The resulting virus mutants were deficient in plaque formation, and titers were reduced more than 100-fold from those of wild-type PrV. Ultrastructural analyses demonstrated that capsid maturation and DNA packaging were not affected. However, neither budding at the inner nuclear membrane nor cytoplasmic or extracellular virus particles were observed. These replication defects were similar to those of a UL34 deletion mutant (B. G. Klupp, H. Granzow, and T. C. Mettenleiter, J. Virol. 74:10063-10073, 2000) and could be completely repaired in a cell line which constitutively expresses the UL31 protein. Yeast two-hybrid studies revealed that a UL31 fusion protein specifically interacts with plasmids of a PrV genome library expressing the N-terminal part of UL34. Vice versa, UL34 selected UL31-encoding plasmids from the library. Immunofluorescence studies and immune electron microscopy demonstrated that in cells infected with wild-type PrV, both proteins accumulate at the nuclear membrane, whereas in the absence of UL34 the UL31 protein is dispersed throughout the nucleus. Like the UL34 protein, the UL31 gene product is a component of enveloped virus particles within the perinuclear space and absent from mature virions. Our findings suggest that physical interaction between these two virus proteins might be a prerequisite for primary envelopment of PrV at the inner nuclear membrane and that this envelope is removed by fusion with the outer nuclear membrane.
Figures


Similar articles
-
Analysis of viral and cellular factors influencing herpesvirus-induced nuclear envelope breakdown.J Virol. 2012 Jun;86(12):6512-21. doi: 10.1128/JVI.00068-12. Epub 2012 Apr 4. J Virol. 2012. PMID: 22491460 Free PMC article.
-
Pseudorabies virus UL37 gene product is involved in secondary envelopment.J Virol. 2001 Oct;75(19):8927-36. doi: 10.1128/JVI.75.19.8927-8936.2001. J Virol. 2001. PMID: 11533156 Free PMC article.
-
Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus.J Gen Virol. 2001 Oct;82(Pt 10):2363-2371. doi: 10.1099/0022-1317-82-10-2363. J Gen Virol. 2001. PMID: 11562530
-
Have NEC Coat, Will Travel: Structural Basis of Membrane Budding During Nuclear Egress in Herpesviruses.Adv Virus Res. 2017;97:107-141. doi: 10.1016/bs.aivir.2016.07.002. Epub 2016 Sep 1. Adv Virus Res. 2017. PMID: 28057257 Free PMC article. Review.
-
Nuclear Egress.Curr Issues Mol Biol. 2021;41:125-170. doi: 10.21775/cimb.041.125. Epub 2020 Aug 7. Curr Issues Mol Biol. 2021. PMID: 32764158 Free PMC article. Review.
Cited by
-
Mechanism of Nuclear Lamina Disruption and the Role of pUS3 in HSV-1 Nuclear Egress.J Virol. 2021 Apr 26;95(10):e02432-20. doi: 10.1128/JVI.02432-20. Epub 2021 Mar 3. J Virol. 2021. PMID: 33658339 Free PMC article.
-
Analysis of viral and cellular factors influencing herpesvirus-induced nuclear envelope breakdown.J Virol. 2012 Jun;86(12):6512-21. doi: 10.1128/JVI.00068-12. Epub 2012 Apr 4. J Virol. 2012. PMID: 22491460 Free PMC article.
-
SUN2 Modulates the Propagation of HSV-1.J Virol. 2022 May 11;96(9):e0045322. doi: 10.1128/jvi.00453-22. Epub 2022 Apr 18. J Virol. 2022. PMID: 35435724 Free PMC article.
-
Intragenic and extragenic suppression of a mutation in herpes simplex virus 1 UL34 that affects both nuclear envelope targeting and membrane budding.J Virol. 2011 Nov;85(22):11615-25. doi: 10.1128/JVI.05730-11. Epub 2011 Sep 7. J Virol. 2011. PMID: 21900173 Free PMC article.
-
Role of tegument proteins in herpesvirus assembly and egress.Protein Cell. 2010 Nov;1(11):987-98. doi: 10.1007/s13238-010-0120-0. Epub 2010 Dec 10. Protein Cell. 2010. PMID: 21153516 Free PMC article. Review.
References
-
- Baer, R., A. T. Bankier, M. D. Biggin, P. L. Deininger, P. J. Farrell, T. J. Gibson, G. Hatfull, G. S. Hudson, S. C. Satchwell, C. Seguin, P. S. Tuffnell, and B. G. Barrell. 1984. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature 310:207–211. - PubMed
-
- Ben Porat, T., J. M. Demarchi, and A. S. Kaplan. 1974. Characterization of defective interfering viral particles present in a population of pseudorabies virions. Virology 61:29–37. - PubMed
-
- Ben Porat, T., R. A. Veach, and S. Ihara. 1983. Localization of the regions of homology between the genomes of herpes simplex virus type 1 and pseudorabies virus. Virology 127:194–204. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

