Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells
- PMID: 11733577
- PMCID: PMC2193527
- DOI: 10.1084/jem.194.11.1625
Gene expression profiling of B cell chronic lymphocytic leukemia reveals a homogeneous phenotype related to memory B cells
Abstract
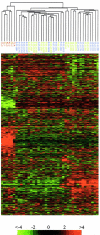
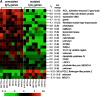
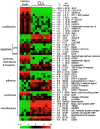
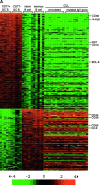
B cell-derived chronic lymphocytic leukemia (B-CLL) represents a common malignancy whose cell derivation and pathogenesis are unknown. Recent studies have shown that >50% of CLLs display hypermutated immunoglobulin variable region (IgV) sequences and a more favorable prognosis, suggesting that they may represent a distinct subset of CLLs which have transited through germinal centers (GCs), the physiologic site of IgV hypermutation. To further investigate the phenotype of CLLs, their cellular derivation and their relationship to normal B cells, we have analyzed their gene expression profiles using oligonucleotide-based DNA chip microarrays representative of approximately 12,000 genes. The results show that CLLs display a common and characteristic gene expression profile that is largely independent of their IgV genotype. Nevertheless, a restricted number of genes (<30) have been identified whose differential expression can distinguish IgV mutated versus unmutated cases and identify them in independent panels of cases. Comparison of CLL profiles with those of purified normal B cell subpopulations indicates that the common CLL profile is more related to memory B cells than to those derived from naive B cells, CD5(+) B cells, and GC centroblasts and centrocytes. Finally, this analysis has identified a subset of genes specifically expressed by CLL cells of potential pathogenetic and clinical relevance.
Figures





Similar articles
-
What do somatic hypermutation and class switch recombination teach us about chronic lymphocytic leukaemia pathogenesis?Curr Top Microbiol Immunol. 2005;294:71-89. doi: 10.1007/3-540-29933-5_5. Curr Top Microbiol Immunol. 2005. PMID: 16323428 Review.
-
Analysis of IgV gene mutations in B cell chronic lymphocytic leukaemia according to antigen-driven selection identifies subgroups with different prognosis and usage of the canonical somatic hypermutation machinery.Br J Haematol. 2004 Jul;126(1):29-42. doi: 10.1111/j.1365-2141.2004.04985.x. Br J Haematol. 2004. PMID: 15198729
-
BCL-6 mutations are associated with immunoglobulin variable heavy chain mutations in B-cell chronic lymphocytic leukemia.Cancer Res. 2000 Oct 15;60(20):5644-8. Cancer Res. 2000. PMID: 11059755
-
New insights into the phenotype and cell derivation of B cell chronic lymphocytic leukemia.Curr Top Microbiol Immunol. 2005;294:31-49. doi: 10.1007/3-540-29933-5_3. Curr Top Microbiol Immunol. 2005. PMID: 16329192 Review.
-
ZAP-70 expression identifies a chronic lymphocytic leukemia subtype with unmutated immunoglobulin genes, inferior clinical outcome, and distinct gene expression profile.Blood. 2003 Jun 15;101(12):4944-51. doi: 10.1182/blood-2002-10-3306. Epub 2003 Feb 20. Blood. 2003. PMID: 12595313
Cited by
-
SF3B1 and other novel cancer genes in chronic lymphocytic leukemia.N Engl J Med. 2011 Dec 29;365(26):2497-506. doi: 10.1056/NEJMoa1109016. Epub 2011 Dec 12. N Engl J Med. 2011. PMID: 22150006 Free PMC article.
-
Post-Transformation IGHV-IGHD-IGHJ Mutations in Chronic Lymphocytic Leukemia B Cells: Implications for Mutational Mechanisms and Impact on Clinical Course.Front Oncol. 2021 May 25;11:640731. doi: 10.3389/fonc.2021.640731. eCollection 2021. Front Oncol. 2021. PMID: 34113563 Free PMC article.
-
Phosphatidylinositol 3-kinase-δ inhibitor CAL-101 shows promising preclinical activity in chronic lymphocytic leukemia by antagonizing intrinsic and extrinsic cellular survival signals.Blood. 2010 Sep 23;116(12):2078-88. doi: 10.1182/blood-2010-02-271171. Epub 2010 Jun 3. Blood. 2010. PMID: 20522708 Free PMC article.
-
AID-induced genotoxic stress promotes B cell differentiation in the germinal center via ATM and LKB1 signaling.Mol Cell. 2010 Sep 24;39(6):873-85. doi: 10.1016/j.molcel.2010.08.019. Mol Cell. 2010. PMID: 20864035 Free PMC article.
-
Surface-antigen expression profiling of B cell chronic lymphocytic leukemia: from the signature of specific disease subsets to the identification of markers with prognostic relevance.J Transl Med. 2006 Mar 1;4:11. doi: 10.1186/1479-5876-4-11. J Transl Med. 2006. PMID: 16509989 Free PMC article.
References
-
- Caligaris-Cappio, F., and T.J. Hamblin. 1999. B-cell chronic lymphocytic leukemia: a bird of a different feather. J. Clin. Oncol. 17:399–408. - PubMed
-
- Döhner, H., S. Stilgenbauer, K. Döhner, M. Bentz, and P. Lichter. 1999. Chromosome aberrations in B-cell chronic lymphocytic leukemia: reassessment based on molecular cytogenetic analysis. J. Mol. Med. 77:266–281. - PubMed
-
- Kipps, T.J. 1998. Chronic lymphocytic leukemia. Curr. Opin. Hematol. 5:244–253. - PubMed
-
- Dighiero, G., T. Kipps, H.W. Schroeder, N. Chiorazzi, F. Stevenson, L.E. Silberstein, F. Caligaris-Cappio, and M. Ferrarini. 1996. What is the CLL B-lymphocyte? Leuk. Lymphoma. 22:13–39. - PubMed
-
- Brezinschek, H.P., S.J. Foster, R.I. Brezinschek, T. Dörner, R. Domiati-Saad, and P.E. Lipsky. 1997. Analysis of the human VH gene repertoire. Differential effects of selection and somatic hypermutation on human peripheral CD5+/IgM+ and CD5−/IgM+ B cells. J. Clin. Invest. 99:2488–2501. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

