Crystal structure of the fission yeast mitochondrial Holliday junction resolvase Ydc2
- PMID: 11726496
- PMCID: PMC125760
- DOI: 10.1093/emboj/20.23.6601
Crystal structure of the fission yeast mitochondrial Holliday junction resolvase Ydc2
Abstract
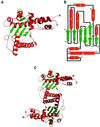
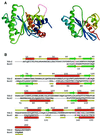
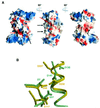
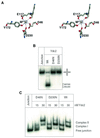
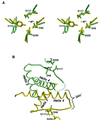
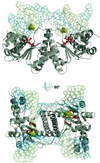
Resolution of Holliday junctions into separate DNA duplexes requires enzymatic cleavage of an equivalent strand from each contributing duplex at or close to the point of strand exchange. Diverse Holliday junction-resolving enzymes have been identified in bacteria, bacteriophages, archaea and pox viruses, but the only eukaryotic examples identified so far are those from fungal mitochondria. We have now determined the crystal structure of Ydc2 (also known as SpCce1), a Holliday junction resolvase from the fission yeast Schizosaccharomyces pombe that is involved in the maintenance of mitochondrial DNA. This first structure of a eukaryotic Holliday junction resolvase confirms a distant evolutionary relationship to the bacterial RuvC family, but reveals structural features which are unique to the eukaryotic enzymes. Detailed analysis of the dimeric structure suggests mechanisms for junction isomerization and communication between the two active sites, and together with site-directed mutagenesis identifies residues involved in catalysis.
Figures

Similar articles
-
Holliday junction resolvase in Schizosaccharomyces pombe has identical endonuclease activity to the CCE1 homologue YDC2.Nucleic Acids Res. 1998 Jan 15;26(2):594-601. doi: 10.1093/nar/26.2.594. Nucleic Acids Res. 1998. PMID: 9421521 Free PMC article.
-
Functional dissection of the Schizosaccharomyces pombe Holliday junction resolvase Ydc2: in vivo role in mitochondrial DNA maintenance.Eur J Biochem. 2003 Jul;270(13):2837-47. doi: 10.1046/j.1432-1033.2003.03661.x. Eur J Biochem. 2003. PMID: 12823554
-
Characterization of a Holliday junction-resolving enzyme from Schizosaccharomyces pombe.Mol Cell Biol. 1997 Nov;17(11):6465-71. doi: 10.1128/MCB.17.11.6465. Mol Cell Biol. 1997. PMID: 9343409 Free PMC article.
-
A new Holliday junction resolving enzyme from Schizosaccharomyces pombe that is homologous to CCE1 from Saccharomyces cerevisiae.J Mol Biol. 1997 Oct 3;272(4):509-22. doi: 10.1006/jmbi.1997.1286. J Mol Biol. 1997. PMID: 9325108
-
The Holliday junction resolvase SpCCE1 prevents mitochondrial DNA aggregation in Schizosaccharomyces pombe.Mol Gen Genet. 2000 Jul;263(6):889-97. doi: 10.1007/s004380000256. Mol Gen Genet. 2000. PMID: 10954073
Cited by
-
The Mrs1 splicing factor binds the bI3 group I intron at each of two tetraloop-receptor motifs.PLoS One. 2010 Feb 1;5(2):e8983. doi: 10.1371/journal.pone.0008983. PLoS One. 2010. PMID: 20126554 Free PMC article.
-
AtGEN1 and AtSEND1, two paralogs in Arabidopsis, possess holliday junction resolvase activity.Plant Physiol. 2014 Sep;166(1):202-16. doi: 10.1104/pp.114.237834. Epub 2014 Jul 18. Plant Physiol. 2014. PMID: 25037209 Free PMC article.
-
Crystal structure of E. coli endonuclease V, an essential enzyme for deamination repair.Sci Rep. 2015 Aug 5;5:12754. doi: 10.1038/srep12754. Sci Rep. 2015. PMID: 26244280 Free PMC article.
-
Mechanism of Transcription Anti-termination in Human Mitochondria.Cell. 2017 Nov 16;171(5):1082-1093.e13. doi: 10.1016/j.cell.2017.09.035. Epub 2017 Oct 12. Cell. 2017. PMID: 29033127 Free PMC article.
-
The search for a human Holliday junction resolvase.Biochem Soc Trans. 2009 Jun;37(Pt 3):519-26. doi: 10.1042/BST0370519. Biochem Soc Trans. 2009. PMID: 19442245 Free PMC article.
References
-
- Ariyoshi M., Vassylyev,D.G., Iwasaki,H., Nakamura,H., Shinagawa,H. and Morikawa,K. (1994) Atomic-structure of the RuvC resolvase—a Holliday junction-specific endonuclease from Escherichia coli. Cell, 78, 1063–1072. - PubMed
-
- Bénard M., Maric,C. and Pierron,G. (2001) DNA replication-dependent formation of joint DNA molecules in Physarum polycephalum. Mol. Cell, 7, 971–980. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

