Binding specificity of Escherichia coli trigger factor
- PMID: 11724963
- PMCID: PMC64667
- DOI: 10.1073/pnas.261432298
Binding specificity of Escherichia coli trigger factor
Abstract
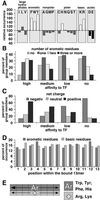
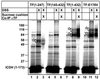
The ribosome-associated chaperone trigger factor (TF) assists the folding of newly synthesized cytosolic proteins in Escherichia coli. Here, we determined the substrate specificity of TF by examining its binding to 2842 membrane-coupled 13meric peptides. The binding motif of TF was identified as a stretch of eight amino acids, enriched in basic and aromatic residues and with a positive net charge. Fluorescence spectroscopy verified that TF exhibited a comparable substrate specificity for peptides in solution. The affinity to peptides in solution was low, indicating that TF requires ribosome association to create high local concentrations of nascent polypeptide substrates for productive interaction in vivo. Binding to membrane-coupled peptides occurred through the central peptidyl-prolyl-cis/trans isomerase (PPIase) domain of TF, however, independently of prolyl residues. Crosslinking experiments showed that a TF fragment containing the PPIase domain linked to the ribosome via the N-terminal domain is sufficient for interaction with nascent polypeptide substrates. Homology modeling of the PPIase domain revealed a conserved FKBP(FK506-binding protein)-like binding pocket composed of exposed aromatic residues embedded in a groove with negative surface charge. The features of this groove complement well the determined substrate specificity of TF. Moreover, a mutation (E178V) in this putative substrate binding groove known to enhance PPIase activity also enhanced TF's association with a prolyl-free model peptide in solution and with nascent polypeptides. This result suggests that both prolyl-independent binding of peptide substrates and peptidyl-prolyl isomerization involve the same binding site.
Figures


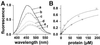
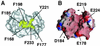
Similar articles
-
Functional dissection of Escherichia coli trigger factor: unraveling the function of individual domains.J Bacteriol. 2004 Jun;186(12):3777-84. doi: 10.1128/JB.186.12.3777-3784.2004. J Bacteriol. 2004. PMID: 15175291 Free PMC article.
-
PPIase domain of trigger factor acts as auxiliary chaperone site to assist the folding of protein substrates bound to the crevice of trigger factor.Int J Biochem Cell Biol. 2010 Jun;42(6):890-901. doi: 10.1016/j.biocel.2010.01.019. Epub 2010 Jan 21. Int J Biochem Cell Biol. 2010. PMID: 20096367
-
Three-state equilibrium of Escherichia coli trigger factor.Biol Chem. 2002 Oct;383(10):1611-9. doi: 10.1515/BC.2002.182. Biol Chem. 2002. PMID: 12452438
-
A cradle for new proteins: trigger factor at the ribosome.Curr Opin Struct Biol. 2005 Apr;15(2):204-12. doi: 10.1016/j.sbi.2005.03.005. Curr Opin Struct Biol. 2005. PMID: 15837180 Review.
-
Peptidyl-prolyl cis-trans isomerases: structure and functions.Biochemistry (Mosc). 1999 Jul;64(7):738-51. Biochemistry (Mosc). 1999. PMID: 10424896 Review.
Cited by
-
Analysis, characterization, and loci of the tuf genes in lactobacillus and bifidobacterium species and their direct application for species identification.Appl Environ Microbiol. 2003 Nov;69(11):6908-22. doi: 10.1128/AEM.69.11.6908-6922.2003. Appl Environ Microbiol. 2003. PMID: 14602655 Free PMC article.
-
Protein folding in vitro and in the cell: From a solitary journey to a team effort.Biophys Chem. 2022 Aug;287:106821. doi: 10.1016/j.bpc.2022.106821. Epub 2022 Apr 29. Biophys Chem. 2022. PMID: 35667131 Free PMC article. Review.
-
Arginine π-stacking drives binding to fibrils of the Alzheimer protein Tau.Nat Commun. 2020 Jan 29;11(1):571. doi: 10.1038/s41467-019-13745-7. Nat Commun. 2020. PMID: 31996674 Free PMC article.
-
Structural features of chloroplast trigger factor determined at 2.6 Å resolution.Acta Crystallogr D Struct Biol. 2022 Oct 1;78(Pt 10):1259-1272. doi: 10.1107/S2059798322009068. Epub 2022 Sep 27. Acta Crystallogr D Struct Biol. 2022. PMID: 36189745 Free PMC article.
-
Ribosome-nascent Chain Interaction Regulates N-terminal Protein Modification.J Mol Biol. 2022 May 15;434(9):167535. doi: 10.1016/j.jmb.2022.167535. Epub 2022 Mar 10. J Mol Biol. 2022. PMID: 35278477 Free PMC article.
References
-
- Bukau B, Deuerling E, Pfund C, Craig E A. Cell. 2000;101:119–122. - PubMed
-
- Ellis R J, Hemmingsen S M. Trends Biochem Sci. 1989;14:339–342. - PubMed
-
- Gething M-J H, Sambrook J F. Nature (London) 1992;355:33–45. - PubMed
-
- Hartl F U. Nature (London) 1996;381:571–580. - PubMed
-
- Horwich A L, Brooks Low K, Fenton W A, Hirshfield I N, Furtak K. Cell. 1993;74:909–917. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

