Organization of two transmissible gastroenteritis coronavirus membrane protein topologies within the virion and core
- PMID: 11711614
- PMCID: PMC116120
- DOI: 10.1128/JVI.75.24.12228-12240.2001
Organization of two transmissible gastroenteritis coronavirus membrane protein topologies within the virion and core
Abstract
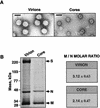
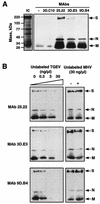
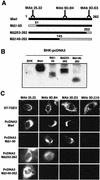
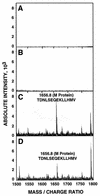
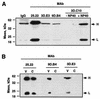
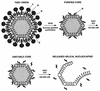
The difference in membrane (M) protein compositions between the transmissible gastroenteritis coronavirus (TGEV) virion and the core has been studied. The TGEV M protein adopts two topologies in the virus envelope, a Nexo-Cendo topology (with the amino terminus exposed to the virus surface and the carboxy terminus inside the virus particle) and a Nexo-Cexo topology (with both the amino and carboxy termini exposed to the virion surface). The existence of a population of M molecules adopting a Nexo-Cexo topology in the virion envelope was demonstrated by (i) immunopurification of (35)S-labeled TGEV virions using monoclonal antibodies (MAbs) specific for the M protein carboxy terminus (this immunopurification was inhibited only by deletion mutant M proteins that maintained an intact carboxy terminus), (ii) direct binding of M-specific MAbs to the virus surface, and (iii) mass spectrometry analysis of peptides released from trypsin-treated virions. Two-thirds of the total number of M protein molecules found in the virion were associated with the cores, and one-third was lost during core purification. MAbs specific for the M protein carboxy terminus were bound to native virions through the M protein in a Nexo-Cexo conformation, and these molecules were removed when the virus envelope was disrupted with NP-40 during virus core purification. All of the M protein was susceptible to N-glycosidase F treatment of the native virions, which indicates that all the M protein molecules are exposed to the virus surface. Cores purified from glycosidase-treated virions included M protein molecules that completely or partially lost the carbohydrate moiety, which strongly suggests that the M protein found in the cores was also exposed in the virus envelope and was not present exclusively in the virus interior. A TGEV virion structure integrating all the data is proposed. According to this working model, the TGEV virion consists of an internal core, made of the nucleocapsid and the carboxy terminus of the M protein, and the envelope, containing the spike (S) protein, the envelope (E) protein, and the M protein in two conformations. The two-thirds of the molecules that are in a Nexo-Cendo conformation (with their carboxy termini embedded within the virus core) interact with the internal core, and the remaining third of the molecules, whose carboxy termini are in a Nexo-Cexo conformation, are lost during virus core purification.
Figures



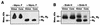
Similar articles
-
The membrane M protein carboxy terminus binds to transmissible gastroenteritis coronavirus core and contributes to core stability.J Virol. 2001 Feb;75(3):1312-24. doi: 10.1128/JVI.75.3.1312-1324.2001. J Virol. 2001. PMID: 11152504 Free PMC article.
-
Membrane protein molecules of transmissible gastroenteritis coronavirus also expose the carboxy-terminal region on the external surface of the virion.J Virol. 1995 Sep;69(9):5269-77. doi: 10.1128/JVI.69.9.5269-5277.1995. J Virol. 1995. PMID: 7636969 Free PMC article.
-
Reconstituted coronavirus TGEV virosomes lose the virus ability to induce porcine interferon-alpha production.Vet Res. 1997;28(1):77-86. Vet Res. 1997. Corrected and republished in: Vet Res. 1997;28(2):105-14.. PMID: 9172843 Corrected and republished.
-
Sialic acid binding activity of transmissible gastroenteritis coronavirus affects sedimentation behavior of virions and solubilized glycoproteins.J Virol. 2001 Jan;75(2):844-9. doi: 10.1128/JVI.75.2.844-849.2001. J Virol. 2001. PMID: 11134297 Free PMC article.
-
Virus particle dynamics.Adv Protein Chem. 2003;64:197-218. doi: 10.1016/s0065-3233(03)01005-2. Adv Protein Chem. 2003. PMID: 13677048 Review. No abstract available.
Cited by
-
mRNA Vaccines against SARS-CoV-2: Advantages and Caveats.Int J Mol Sci. 2023 Mar 21;24(6):5944. doi: 10.3390/ijms24065944. Int J Mol Sci. 2023. PMID: 36983017 Free PMC article. Review.
-
Porcine Epidemic Diarrhea Virus: An Updated Overview of Virus Epidemiology, Virulence Variation Patterns and Virus-Host Interactions.Viruses. 2022 Nov 2;14(11):2434. doi: 10.3390/v14112434. Viruses. 2022. PMID: 36366532 Free PMC article. Review.
-
Immune Profiling Uncovers Memory T-Cell Responses with a Th17 Signature in Cancer Patients with Previous SARS-CoV-2 Infection Followed by mRNA Vaccination.Cancers (Basel). 2022 Sep 14;14(18):4464. doi: 10.3390/cancers14184464. Cancers (Basel). 2022. PMID: 36139625 Free PMC article.
-
Development of a SARS-CoV-2 nucleocapsid specific monoclonal antibody.Virology. 2021 Jun;558:28-37. doi: 10.1016/j.virol.2021.01.003. Epub 2021 Feb 1. Virology. 2021. PMID: 33714753 Free PMC article.
-
The human pandemic coronaviruses on the show: The spike glycoprotein as the main actor in the coronaviruses play.Int J Biol Macromol. 2021 May 15;179:1-19. doi: 10.1016/j.ijbiomac.2021.02.203. Epub 2021 Mar 2. Int J Biol Macromol. 2021. PMID: 33667553 Free PMC article. Review.
References
-
- Ansorge W. Fast and sensitive detection of protein and DNA bands by treatment with potassium permanganate. J Biochem Biophys Methods. 1985;11:13–20. - PubMed
-
- Cavanagh D. Coronavirus IBV glycopolypeptides: size of their polypeptide moieties and nature of their oligosaccharides. J Gen Virol. 1983;64:1187–1191. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

