IL-1 induces collagenase-3 (MMP-13) promoter activity in stably transfected chondrocytic cells: requirement for Runx-2 and activation by p38 MAPK and JNK pathways
- PMID: 11691923
- PMCID: PMC60184
- DOI: 10.1093/nar/29.21.4361
IL-1 induces collagenase-3 (MMP-13) promoter activity in stably transfected chondrocytic cells: requirement for Runx-2 and activation by p38 MAPK and JNK pathways
Abstract
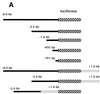
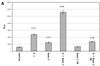
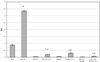
Osteoarthritic chondrocytes secrete matrix metalloproteinase-13 (MMP-13) in response to interleukin-1 (IL-1), causing digestion of type II collagen in cartilage. Using chondrocytic cells, we previously determined that IL-1 induced a strong MMP-13 transcriptional response that requires p38 MAPK, JNK and the transcription factor NF-kappaB. Now, we have studied the tissue-specific transcriptional regulation of MMP-13. Constitutive expression of the transcription factor Runx-2 correlated with the ability of a cell type to express MMP-13 and was required for IL-1 induction; moreover, Runx-2 enhanced IL-1 induction of MMP-13 transcription by synergizing with the p38 MAPK signaling pathway. Transiently transfected MMP-13 promoters were not IL-1 inducible. However, -405 bp of stably integrated promoter was sufficient for 5- to 6-fold IL-1 induction of reporter activity and this integrated reporter required the same p38 MAPK pathway as the endogenous gene. Finally, mutation of the proximal Runx binding site and the proximal AP-1 site blunted the transcriptional response to IL-1, and double mutation synergistically decreased reporter activity. In summary, our data suggest that the transcriptional MMP-13 response to IL-1 is controlled by the p38 pathway interacting at the MMP-13 promoter through the tissue-specific transcription factor Runx-2 and the ubiquitous AP-1 transcription factor.
Figures










Similar articles
-
Interleukin-1 induction of collagenase 3 (matrix metalloproteinase 13) gene expression in chondrocytes requires p38, c-Jun N-terminal kinase, and nuclear factor kappaB: differential regulation of collagenase 1 and collagenase 3.Arthritis Rheum. 2000 Apr;43(4):801-11. doi: 10.1002/1529-0131(200004)43:4<801::AID-ANR10>3.0.CO;2-4. Arthritis Rheum. 2000. PMID: 10765924
-
Inhibition of interleukin-1-stimulated MAP kinases, activating protein-1 (AP-1) and nuclear factor kappa B (NF-kappa B) transcription factors down-regulates matrix metalloproteinase gene expression in articular chondrocytes.Matrix Biol. 2002 Apr;21(3):251-62. doi: 10.1016/s0945-053x(02)00007-0. Matrix Biol. 2002. PMID: 12009331
-
Amplification of IL-1 beta-induced matrix metalloproteinase-9 expression by superoxide in rat glomerular mesangial cells is mediated by increased activities of NF-kappa B and activating protein-1 and involves activation of the mitogen-activated protein kinase pathways.J Immunol. 2000 Nov 15;165(10):5788-97. doi: 10.4049/jimmunol.165.10.5788. J Immunol. 2000. PMID: 11067938
-
Transcriptional regulation of collagenase (MMP-1, MMP-13) genes in arthritis: integration of complex signaling pathways for the recruitment of gene-specific transcription factors.Arthritis Res. 2002;4(3):157-64. doi: 10.1186/ar401. Epub 2001 Nov 23. Arthritis Res. 2002. PMID: 12010565 Free PMC article. Review.
-
Stress signaling in Drosophila.Oncogene. 1999 Nov 1;18(45):6172-82. doi: 10.1038/sj.onc.1203125. Oncogene. 1999. PMID: 10557109 Review.
Cited by
-
4-Methylumbelliferone Diminishes Catabolically Activated Articular Chondrocytes and Cartilage Explants via a Mechanism Independent of Hyaluronan Inhibition.J Biol Chem. 2016 Jun 3;291(23):12087-104. doi: 10.1074/jbc.M115.709683. Epub 2016 Apr 25. J Biol Chem. 2016. PMID: 27129266 Free PMC article.
-
CXCL12/CXCR4 Axis Regulates Aggrecanase Activation and Cartilage Degradation in a Post-Traumatic Osteoarthritis Rat Model.Int J Mol Sci. 2016 Sep 29;17(10):1522. doi: 10.3390/ijms17101522. Int J Mol Sci. 2016. PMID: 27690009 Free PMC article.
-
Essential role for c-Jun N-terminal kinase 2 in corneal epithelial response to desiccating stress.Arch Ophthalmol. 2009 Dec;127(12):1625-31. doi: 10.1001/archophthalmol.2009.316. Arch Ophthalmol. 2009. PMID: 20008718 Free PMC article.
-
Cytokine-induced cysteine- serine-rich nuclear protein-1 (CSRNP1) selectively contributes to MMP1 expression in human chondrocytes.PLoS One. 2018 Nov 15;13(11):e0207240. doi: 10.1371/journal.pone.0207240. eCollection 2018. PLoS One. 2018. PMID: 30440036 Free PMC article.
-
Mmp13 deletion in mesenchymal cells increases bone mass and may attenuate the cortical bone loss caused by estrogen deficiency.Sci Rep. 2022 Jun 17;12(1):10257. doi: 10.1038/s41598-022-14470-w. Sci Rep. 2022. PMID: 35715555 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

