Reconstitution of recombinant chromatin establishes a requirement for histone-tail modifications during chromatin assembly and transcription
- PMID: 11691835
- PMCID: PMC312801
- DOI: 10.1101/gad.937401
Reconstitution of recombinant chromatin establishes a requirement for histone-tail modifications during chromatin assembly and transcription
Abstract
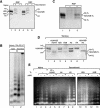
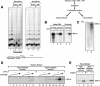
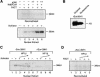
The human ISWI-containing factor RSF (remodeling and spacing factor) was found to mediate nucleosome deposition and, in the presence of ATP, generate regularly spaced nucleosome arrays. Using this system, recombinant chromatin was reconstituted with bacterially produced histones. Acetylation of the histone tails was found to play an important role in establishing regularly spaced nucleosome arrays. Recombinant chromatin lacking histone acetylation was impaired in directing transcription. Histone-tail modifications were found to regulate transcription from the recombinant chromatin. Acetylation of the histone tails by p300 was found to increase transcription. Methylation of the histone H3 tail by Suv39H1 was found to repress transcription in an HP1-dependent manner. The effects of histone-tail modifications were observed in nuclear extracts. A highly reconstituted RNA polymerase II transcription system was refractory to the effect imposed by acetylation and methylation.
Figures



Similar articles
-
p300-mediated acetylation facilitates the transfer of histone H2A-H2B dimers from nucleosomes to a histone chaperone.Genes Dev. 2000 Aug 1;14(15):1899-907. Genes Dev. 2000. PMID: 10921904 Free PMC article.
-
Histone H3 lysine 4 methylation disrupts binding of nucleosome remodeling and deacetylase (NuRD) repressor complex.J Biol Chem. 2002 Apr 5;277(14):11621-4. doi: 10.1074/jbc.C200045200. Epub 2002 Feb 15. J Biol Chem. 2002. PMID: 11850414
-
Direct binding of INHAT to H3 tails disrupted by modifications.J Biol Chem. 2004 Jun 4;279(23):23859-62. doi: 10.1074/jbc.C400151200. Epub 2004 Apr 19. J Biol Chem. 2004. PMID: 15100215
-
Secondary structures of the core histone N-terminal tails: their role in regulating chromatin structure.Subcell Biochem. 2013;61:37-55. doi: 10.1007/978-94-007-4525-4_2. Subcell Biochem. 2013. PMID: 23150245 Review.
-
[Histone variants and histone exchange].Yi Chuan. 2006 Apr;28(4):493-500. Yi Chuan. 2006. PMID: 16606605 Review. Chinese.
Cited by
-
Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo.Genes Dev. 2004 Jan 15;18(2):170-83. doi: 10.1101/gad.1139604. Genes Dev. 2004. PMID: 14752009 Free PMC article.
-
Functional proteomics establishes the interaction of SIRT7 with chromatin remodeling complexes and expands its role in regulation of RNA polymerase I transcription.Mol Cell Proteomics. 2012 May;11(5):60-76. doi: 10.1074/mcp.A111.015156. Mol Cell Proteomics. 2012. PMID: 22586326 Free PMC article.
-
Identification and characterization of ToRC, a novel ISWI-containing ATP-dependent chromatin assembly complex.Genes Dev. 2012 Mar 15;26(6):603-14. doi: 10.1101/gad.180604.111. Genes Dev. 2012. PMID: 22426536 Free PMC article.
-
Facile synthesis of site-specifically acetylated and methylated histone proteins: reagents for evaluation of the histone code hypothesis.Proc Natl Acad Sci U S A. 2003 Oct 14;100(21):12033-8. doi: 10.1073/pnas.2035256100. Epub 2003 Oct 6. Proc Natl Acad Sci U S A. 2003. PMID: 14530408 Free PMC article.
-
ISWI chromatin remodeling: one primary actor or a coordinated effort?Curr Opin Struct Biol. 2014 Feb;24:150-5. doi: 10.1016/j.sbi.2014.01.010. Epub 2014 Feb 19. Curr Opin Struct Biol. 2014. PMID: 24561830 Free PMC article. Review.
References
-
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell. 2000;103:667–678. - PubMed
-
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. - PubMed
-
- Berger SL. Molecular biology: The histone modification circus. Science. 2001;292:64–65. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
