Two checkpoint complexes are independently recruited to sites of DNA damage in vivo
- PMID: 11691833
- PMCID: PMC312815
- DOI: 10.1101/gad.903501
Two checkpoint complexes are independently recruited to sites of DNA damage in vivo
Abstract
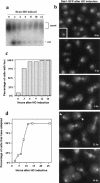
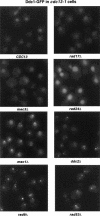
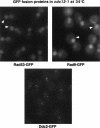
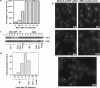
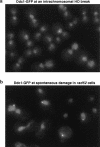
The Ddc1/Rad17/Mec3 complex and Rad24 are DNA damage checkpoint components with limited homology to replication factors PCNA and RF-C, respectively, suggesting that these factors promote checkpoint activation by "sensing" DNA damage directly. Mec1 kinase, however, phosphorylates the checkpoint protein Ddc2 in response to damage in the absence of all other known checkpoint proteins, suggesting instead that Mec1 and/or Ddc2 may act as the initial sensors of DNA damage. In this paper, we show that Ddc1 or Ddc2 fused to GFP localizes to a single subnuclear focus following an endonucleolytic break. Other forms of damage result in a greater number of Ddc1-GFP or Ddc2-GFP foci, in correlation with the number of damage sites generated, indicating that Ddc1 and Ddc2 are both recruited to sites of DNA damage. Interestingly, Ddc2 localization is severely abrogated in mec1 cells but requires no other known checkpoint genes, whereas Ddc1 localization requires Rad17, Mec3, and Rad24, but not Mec1. Therefore, Ddc1 and Ddc2 recognize DNA damage by independent mechanisms. These data support a model in which assembly of multiple checkpoint complexes at DNA damage sites stimulates checkpoint activation. Further, we show that although Ddc1 remains strongly localized following checkpoint adaptation, many nuclei contain only dim foci of Ddc2-GFP, suggesting that Ddc2 localization may be down-regulated during resumption of cell division. Lastly, visualization of checkpoint proteins localized to damage sites serves as a useful tool for analysis of DNA damage in living cells.
Figures


Similar articles
-
The checkpoint clamp activates Mec1 kinase during initiation of the DNA damage checkpoint.Mol Cell. 2006 Dec 28;24(6):891-901. doi: 10.1016/j.molcel.2006.11.027. Mol Cell. 2006. PMID: 17189191 Free PMC article.
-
Ddc2ATRIP promotes Mec1ATR activation at RPA-ssDNA tracts.PLoS Genet. 2019 Aug 1;15(8):e1008294. doi: 10.1371/journal.pgen.1008294. eCollection 2019 Aug. PLoS Genet. 2019. PMID: 31369547 Free PMC article.
-
Ddc2 mediates Mec1 activation through a Ddc1- or Dpb11-independent mechanism.PLoS Genet. 2014 Feb 20;10(2):e1004136. doi: 10.1371/journal.pgen.1004136. eCollection 2014 Feb. PLoS Genet. 2014. PMID: 24586187 Free PMC article.
-
Cell-cycle-specific activators of the Mec1/ATR checkpoint kinase.Biochem Soc Trans. 2011 Apr;39(2):600-5. doi: 10.1042/BST0390600. Biochem Soc Trans. 2011. PMID: 21428947 Review.
-
Activation of ATR-related protein kinase upon DNA damage recognition.Curr Genet. 2020 Apr;66(2):327-333. doi: 10.1007/s00294-019-01039-w. Epub 2019 Oct 17. Curr Genet. 2020. PMID: 31624858 Free PMC article. Review.
Cited by
-
Chk2 homolog Mek1 limits exonuclease 1-dependent DNA end resection during meiotic recombination in Saccharomyces cerevisiae.Genetics. 2024 Sep 4;228(1):iyae112. doi: 10.1093/genetics/iyae112. Genetics. 2024. PMID: 39005070
-
DNA damage checkpoint triggers autophagy to regulate the initiation of anaphase.Proc Natl Acad Sci U S A. 2013 Jan 2;110(1):E41-9. doi: 10.1073/pnas.1218065109. Epub 2012 Nov 19. Proc Natl Acad Sci U S A. 2013. PMID: 23169651 Free PMC article.
-
An N-terminal acidic region of Sgs1 interacts with Rpa70 and recruits Rad53 kinase to stalled forks.EMBO J. 2012 Sep 12;31(18):3768-83. doi: 10.1038/emboj.2012.195. Epub 2012 Jul 20. EMBO J. 2012. PMID: 22820947 Free PMC article.
-
Association of Rad9 with double-strand breaks through a Mec1-dependent mechanism.Mol Cell Biol. 2004 Apr;24(8):3277-85. doi: 10.1128/MCB.24.8.3277-3285.2004. Mol Cell Biol. 2004. PMID: 15060150 Free PMC article.
-
A requirement for replication in activation of the ATR-dependent DNA damage checkpoint.Genes Dev. 2002 Sep 15;16(18):2327-32. doi: 10.1101/gad.1013502. Genes Dev. 2002. PMID: 12231621 Free PMC article.
References
-
- Burtelow MA, Kaufmann SH, Karnitz LM. Retention of the human Rad9 checkpoint complex in extraction-resistant nuclear complexes after DNA damage. J Biol Chem. 2000;275:26343–26348. - PubMed
-
- Edwards RJ, Bentley RJ, Carr AM. A Rad3–Rad26 complex responds to DNA damage independently of other checkpoint proteins. Nat Cell Biol. 1999;1:393–398. - PubMed
-
- Emili A. MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol Cell. 1998;2:183–189. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
