Murine leukemia virus proviral insertions between the N-ras and unr genes in B-cell lymphoma DNA affect the expression of N-ras only
- PMID: 11689675
- PMCID: PMC114780
- DOI: 10.1128/JVI.75.23.11907-11912.2001
Murine leukemia virus proviral insertions between the N-ras and unr genes in B-cell lymphoma DNA affect the expression of N-ras only
Abstract
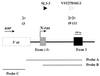
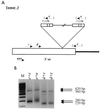
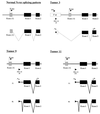
Akv1-99, a variant of Akv murine leukemia virus, induces B-cell lymphomas with nearly 100% incidence and a mean latency period of 12 months after injection into newborn NMRI mice. PCR amplification and sequence analyses of DNA flanking integrated proviruses revealed proviral insertion into the N-ras/unr (upstream of N-ras) locus in 2 out of 13 B-cell lymphomas, both of which appeared clonal by Southern blotting analysis. These two tumors showed increased expression levels of N-ras by Northern blotting, as did a third tumor shown by reverse transcriptase PCR to have a nonclonal provirus integration located in the same area. However, no significant changes in expression were observed when using a specific probe for the unr gene. All proviruses were integrated in the same transcriptional orientation as unr and N-ras genes. By promoter insertion, the two Akv1-99 proviruses integrated between exon -1 and exon 1 of N-ras gave rise to two different spliced products, whereas the provirus integrated into unr used only an exon skipping pattern. The absence of mutations of the N-ras codons 12, 13, 18, and 61 suggests that activation of the proto-oncogene is exclusively due to overexpression by retroviral promoter insertion, and furthermore, Northern blot analyses indicate that the expression of unr is unaffected by N-ras overexpression even in the case where the unr gene itself is the target of proviral insertion. Thus, altogether our findings indicate that overexpression of N-ras plays a role in development of murine leukemia virus-induced B-cell lymphomas, leaving the expression of the tightly linked unr gene unaltered.
Figures


Similar articles
-
Tumor model-specific proviral insertional mutagenesis of the Fos/Jdp2/Batf locus.Virology. 2005 Jul 5;337(2):353-64. doi: 10.1016/j.virol.2005.04.027. Virology. 2005. PMID: 15913695
-
Enhancer mutations of Akv murine leukemia virus inhibit the induction of mature B-cell lymphomas and shift disease specificity towards the more differentiated plasma cell stage.Virology. 2007 May 25;362(1):179-91. doi: 10.1016/j.virol.2006.12.016. Epub 2007 Jan 29. Virology. 2007. PMID: 17258785
-
Non-identical patterns of proviral insertions around host transcription units in lymphomas induced by different strains of murine leukemia virus.Virology. 2006 Sep 15;353(1):193-9. doi: 10.1016/j.virol.2006.05.024. Epub 2006 Jun 21. Virology. 2006. PMID: 16790256
-
Ets and retroviruses - transduction and activation of members of the Ets oncogene family in viral oncogenesis.Oncogene. 2000 Dec 18;19(55):6472-81. doi: 10.1038/sj.onc.1204046. Oncogene. 2000. PMID: 11175363 Review.
-
Regulation of the expression and activity of Unr in mammalian cells.Biochem Soc Trans. 2015 Dec;43(6):1241-6. doi: 10.1042/BST20150165. Biochem Soc Trans. 2015. PMID: 26614667 Review.
Cited by
-
Nras overexpression results in granulocytosis, T-cell expansion and early lethality in mice.PLoS One. 2012;7(8):e42216. doi: 10.1371/journal.pone.0042216. Epub 2012 Aug 2. PLoS One. 2012. PMID: 22876308 Free PMC article.
-
Forward and Reverse Genetics of B Cell Malignancies: From Insertional Mutagenesis to CRISPR-Cas.Front Immunol. 2021 Aug 13;12:670280. doi: 10.3389/fimmu.2021.670280. eCollection 2021. Front Immunol. 2021. PMID: 34484175 Free PMC article. Review.
-
UNR/CSDE1 Expression Is Critical to Maintain Invasive Phenotype of Colorectal Cancer through Regulation of c-MYC and Epithelial-to-Mesenchymal Transition.J Clin Med. 2019 Apr 25;8(4):560. doi: 10.3390/jcm8040560. J Clin Med. 2019. PMID: 31027221 Free PMC article.
-
Deregulated Nras expression in knock-in animals harboring a gammaretroviral long terminal repeat at the Nras/Csde1 locus.PLoS One. 2013;8(2):e56029. doi: 10.1371/journal.pone.0056029. Epub 2013 Feb 13. PLoS One. 2013. PMID: 23418499 Free PMC article.
-
Radiation leukemia virus common integration at the Kis2 locus: simultaneous overexpression of a novel noncoding RNA and of the proximal Phf6 gene.J Virol. 2005 Sep;79(17):11443-56. doi: 10.1128/JVI.79.17.11443-11456.2005. J Virol. 2005. PMID: 16103195 Free PMC article.
References
-
- Barbacid M. ras genes. Annu Rev Biochem. 1987;56:779–827. - PubMed
-
- Bos J L. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. . (Erratum, 50:1352, 1990.) - PubMed
-
- Boussadia O, Amiot F, Cases S, Triqueneaux G, Jacquemin-Sablon H, Dautry F. Transcription of unr (upstream of N-ras) down-modulates N-ras expression in vivo. FEBS Lett. 1997;420:20–24. - PubMed
-
- Boussadia O, Jacquemin-Sablon H, Dautry F. Exon skipping in the expression of the gene immediately upstream of N-ras (unr/NRU) Biochim Biophys Acta. 1993;1172:64–72. - PubMed
-
- Chang H Y, Guerrero I, Lake R, Pellicer A, D'Eustachio P. Mouse N-ras genes: organization of the functional locus and of a truncated cDNA-like pseudogene. Oncogene Res. 1987;1:129–136. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

