The crystal structures of glutathione S-transferases isozymes 1-3 and 1-4 from Anopheles dirus species B
- PMID: 11604524
- PMCID: PMC2374065
- DOI: 10.1110/ps.ps.21201
The crystal structures of glutathione S-transferases isozymes 1-3 and 1-4 from Anopheles dirus species B
Abstract
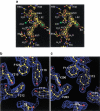
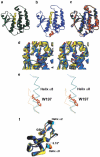
Glutathione S-transferases (GSTs) are dimeric proteins that play an important role in cellular detoxification. Four GSTs from the mosquito Anopheles dirus species B (Ad), an important malaria vector in South East Asia, are produced by alternate splicing of a single transcription product and were previously shown to have detoxifying activity towards pesticides such as DDT. We have determined the crystal structures for two of these alternatively spliced proteins, AdGST1-3 (complexed with glutathione) and AdGST1-4 (apo form), at 1.75 and 2.45 A resolution, respectively. These GST isozymes show differences from the related GST from the Australian sheep blowfly Lucilia cuprina; in particular, the presence of a C-terminal helix forming part of the active site. This helix causes the active site of the Anopheles GSTs to be enclosed. The glutathione-binding helix alpha2 and flanking residues are disordered in the AdGST1-4 (apo) structure, yet ordered in the AdGST1-3 (GSH-bound) structure, suggesting that insect GSTs operate with an induced fit mechanism similar to that found in the plant phi- and human pi-class GSTs. Despite the high overall sequence identities, the active site residues of AdGST1-4 and AdGST1-3 have different conformations.
Figures

Similar articles
-
Catalytic and structural contributions for glutathione-binding residues in a Delta class glutathione S-transferase.Biochem J. 2004 Sep 1;382(Pt 2):751-7. doi: 10.1042/BJ20040697. Biochem J. 2004. PMID: 15182230 Free PMC article.
-
Heterologous expression and characterization of alternatively spliced glutathione S-transferases from a single Anopheles gene.Insect Biochem Mol Biol. 2001 Jul 26;31(9):867-75. doi: 10.1016/s0965-1748(01)00032-7. Insect Biochem Mol Biol. 2001. PMID: 11439246
-
Crystallization of two glutathione S-transferases from an unusual gene family.Acta Crystallogr D Biol Crystallogr. 2001 Jun;57(Pt 6):870-2. doi: 10.1107/s0907444901004929. Epub 2001 May 25. Acta Crystallogr D Biol Crystallogr. 2001. PMID: 11375512
-
Alternative splicing of glutathione S-transferases.Methods Enzymol. 2005;401:100-16. doi: 10.1016/S0076-6879(05)01006-2. Methods Enzymol. 2005. PMID: 16399381 Review.
-
Mosquito glutathione transferases.Methods Enzymol. 2005;401:226-41. doi: 10.1016/S0076-6879(05)01014-1. Methods Enzymol. 2005. PMID: 16399389 Review.
Cited by
-
Expression Patterns of Drosophila Melanogaster Glutathione Transferases.Insects. 2022 Jul 7;13(7):612. doi: 10.3390/insects13070612. Insects. 2022. PMID: 35886788 Free PMC article.
-
Monomeric Camelus dromedarius GSTM1 at low pH is structurally more thermostable than its native dimeric form.PLoS One. 2018 Oct 10;13(10):e0205274. doi: 10.1371/journal.pone.0205274. eCollection 2018. PLoS One. 2018. PMID: 30303997 Free PMC article.
-
Structural and Thermodynamic Insights into Dimerization Interfaces of Drosophila Glutathione Transferases.Biomolecules. 2024 Jun 26;14(7):758. doi: 10.3390/biom14070758. Biomolecules. 2024. PMID: 39062472 Free PMC article.
-
Catalytic and structural contributions for glutathione-binding residues in a Delta class glutathione S-transferase.Biochem J. 2004 Sep 1;382(Pt 2):751-7. doi: 10.1042/BJ20040697. Biochem J. 2004. PMID: 15182230 Free PMC article.
-
Investigating homology between proteins using energetic profiles.PLoS Comput Biol. 2010 Mar 26;6(3):e1000722. doi: 10.1371/journal.pcbi.1000722. PLoS Comput Biol. 2010. PMID: 20361049 Free PMC article.
References
-
- Board, P.G., Coggan, M., Chelvanayagam, G., Easteal, S., Jermiin, L.S., Schulte, G.K., Danley, D.E., Hoth, L.R., Griffor, M.C., Kamath, et al. 2000. Ideication, characterization, and crystal structure of the omega class glutathione transferases. J. Biol. Chem. 275 24798–24806. - PubMed
-
- Brünger, A.T., Adams, P.D., Clore, G.M., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.-S., Kuszewski, J., Nilges, N., Pannu, N.S., Read, et al. 1998. Crystallography and NMR system (CNS): A new software system for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54 905–921. - PubMed
-
- Caccuri, A.M., Antonini, G., Nicotra, M., Battistoni, A., Bello, M.L., Board, P.G., Parker, M.W., and Ricci, G. 1997. Catalytic mechanism and role of hydroxyl residues in the active site of theta class glutathione S-transferases. Investigation of Ser-9 and Tyr-113 in a glutathione S-transferase from the Australian sheep blowfly, Lucilia cuprina. J. Biol. Chem. 272 29681–29686. - PubMed
-
- Cameron, A.D., Sinning, I., L'Hermite, G., Olin, B., Board, P.G., Mannervik, B., and Jones, T.A. 1995. Structural analysis of human alpha-class glutathione transferase A1–1 in the apo-form and in complexes with ethacrynic acid and its glutathione conjugate. Structure 3 717–727. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

