Kinetics of human immunodeficiency virus type 1 (HIV) DNA integration in acutely infected cells as determined using a novel assay for detection of integrated HIV DNA
- PMID: 11602768
- PMCID: PMC114708
- DOI: 10.1128/JVI.75.22.11253-11260.2001
Kinetics of human immunodeficiency virus type 1 (HIV) DNA integration in acutely infected cells as determined using a novel assay for detection of integrated HIV DNA
Abstract
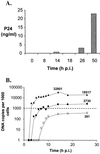
We have developed a novel linker-primer PCR assay for the detection and quantification of integrated human immunodeficiency virus type 1 (HIV) DNA. This assay reproducibly allowed the detection of 10 copies of integrated HIV DNA, in a background of 2 x 10(5) cell equivalents of human chromosomal DNA, without amplifying extrachromosomal HIV DNA. We have used this assay and a near-synchronous one-step T-cell infection model to investigate the kinetics of viral DNA accumulation following HIV infection. We report here that integrated HIV DNA started accumulating 1 h after the first appearance of extrachromosomal viral DNA and accounted for approximately 10% of the total HIV DNA synthesized in the first round of viral replication. These results highlight the efficient nature of integrase-mediated HIV integration in infected T cells.
Figures


Similar articles
-
Analysis of human immunodeficiency virus type 1 integration by using a specific, sensitive and quantitative assay based on real-time polymerase chain reaction.Virus Genes. 2006 Feb;32(1):105-13. doi: 10.1007/s11262-005-5851-2. Virus Genes. 2006. PMID: 16525741
-
Dual phasic suppression of viral replication following de novo human immunodeficiency virus type 1 (HIV-1) infection in lymphocytes of asymptomatic HIV-1 carriers.Leukemia. 1997 Apr;11 Suppl 3:545-7. Leukemia. 1997. PMID: 9209452
-
Self-limiting, cell type-dependent replication of an integrase-defective human immunodeficiency virus type 1 in human primary macrophages but not T lymphocytes.Virology. 1995 Apr 1;208(1):242-8. doi: 10.1006/viro.1995.1148. Virology. 1995. PMID: 11831706
-
Alu-LTR real-time nested PCR assay for quantifying integrated HIV-1 DNA.Methods Mol Biol. 2005;304:139-54. doi: 10.1385/1-59259-907-9:139. Methods Mol Biol. 2005. PMID: 16061972
-
Altered replication of human immunodeficiency virus type 1 (HIV-1) in T cell lines retrovirally transduced to express Herpesvirus saimiri proteins StpC and/or Tip.Virology. 1999 Nov 10;264(1):125-33. doi: 10.1006/viro.1999.9988. Virology. 1999. PMID: 10544137
Cited by
-
Towards an HIV-1 cure: measuring the latent reservoir.Trends Microbiol. 2015 Apr;23(4):192-203. doi: 10.1016/j.tim.2015.01.013. Epub 2015 Mar 5. Trends Microbiol. 2015. PMID: 25747663 Free PMC article. Review.
-
The Molecular Characterization of Intestinal Explant HIV Infection Using Polymerase Chain Reaction-Based Techniques.AIDS Res Hum Retroviruses. 2015 Oct;31(10):981-91. doi: 10.1089/AID.2015.0165. Epub 2015 Aug 24. AIDS Res Hum Retroviruses. 2015. PMID: 26214703 Free PMC article.
-
Human immunodeficiency virus type 1 Vpu and cellular TASK proteins suppress transcription of unintegrated HIV-1 DNA.Virol J. 2012 Nov 19;9:277. doi: 10.1186/1743-422X-9-277. Virol J. 2012. PMID: 23164059 Free PMC article.
-
The largest HIV-1-infected T cell clones in children on long-term combination antiretroviral therapy contain solo LTRs.mBio. 2023 Aug 31;14(4):e0111623. doi: 10.1128/mbio.01116-23. Epub 2023 Aug 2. mBio. 2023. PMID: 37530525 Free PMC article.
-
SAMHD1 restricts HIV-1 replication and regulates interferon production in mouse myeloid cells.PLoS One. 2014 Feb 19;9(2):e89558. doi: 10.1371/journal.pone.0089558. eCollection 2014. PLoS One. 2014. PMID: 24586870 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

