SUMO modification of Rad22, the Schizosaccharomyces pombe homologue of the recombination protein Rad52
- PMID: 11600706
- PMCID: PMC60211
- DOI: 10.1093/nar/29.20.4179
SUMO modification of Rad22, the Schizosaccharomyces pombe homologue of the recombination protein Rad52
Abstract
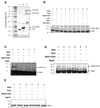
The Schizosaccharomyces pombe rad31 and hus5 genes are required for the DNA damage response, as mutants defective in these genes are sensitive to DNA damaging agents, such as UV and ionising radiation and to the DNA synthesis inhibitor hydroxyurea (HU). Sequence analysis has suggested that rad31 and hus5 encode components of the Pmt3 (SUMO) modification process in S.pombe. We show here that the rad31 null and hus5.62 mutants display reduced levels of Pmt3 modification. We have initiated a search for proteins required for the DNA damage response, which may be modified by Pmt3 and have identified Rad22, the fission yeast homologue of the recombination protein Rad52. Purification of myc + His-tagged Rad22 protein from cells expressing HA-tagged Pmt3 identifies an 83 kDa species which cross-reacts with anti-HA antisera. We show here that Rad22 interacts with Rhp51 and Rpa70 (the fission yeast homologues of Rad51 and the large subunit of RPA, respectively), but that neither of these proteins appears to be responsible for the 83 kDa species. The 83 kDa species is observed when extracts are prepared under both native and denaturing conditions, and is also observed when myc + His-tagged Rad22 and Pmt3 are expressed at wild type levels, suggesting that Rad22 is modified by Pmt3 in vivo. We have established an S.pombe in vitro Pmt3 modification system and have shown that Rad22 and Rhp51 are modified in vitro, but that Rpa70 is not.
Figures



Similar articles
-
Characterization of SUMO-conjugating enzyme mutants in Schizosaccharomyces pombe identifies a dominant-negative allele that severely reduces SUMO conjugation.Biochem J. 2003 May 15;372(Pt 1):97-104. doi: 10.1042/BJ20021645. Biochem J. 2003. PMID: 12597774 Free PMC article.
-
Physical interaction between recombinational proteins Rhp51 and Rad22 in Schizosaccharomyces pombe.J Biol Chem. 2002 Aug 16;277(33):30264-70. doi: 10.1074/jbc.M202517200. Epub 2002 Jun 5. J Biol Chem. 2002. PMID: 12050150
-
Cell-cycle-dependent localisation of Ulp1, a Schizosaccharomyces pombe Pmt3 (SUMO)-specific protease.J Cell Sci. 2002 Mar 15;115(Pt 6):1113-22. doi: 10.1242/jcs.115.6.1113. J Cell Sci. 2002. PMID: 11884512
-
Molecular biology of DNA repair in the fission yeast Schizosaccharomyces pombe.Mutat Res. 1996 Aug 8;363(3):147-61. doi: 10.1016/0921-8777(96)00017-1. Mutat Res. 1996. PMID: 8765156 Review. No abstract available.
-
Repair of UV damage in the fission yeast Schizosaccharomyces pombe.Mutat Res. 2000 Jun 30;451(1-2):197-210. doi: 10.1016/s0027-5107(00)00050-6. Mutat Res. 2000. PMID: 10915873 Review.
Cited by
-
Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins.EMBO J. 2007 Sep 19;26(18):4102-12. doi: 10.1038/sj.emboj.7601839. Epub 2007 Aug 30. EMBO J. 2007. PMID: 17762864 Free PMC article.
-
Nep1, a Schizosaccharomyces pombe deneddylating enzyme.Biochem J. 2005 Jul 15;389(Pt 2):307-14. doi: 10.1042/BJ20041991. Biochem J. 2005. PMID: 15769255 Free PMC article.
-
Chromatin PTEN is involved in DNA damage response partly through regulating Rad52 sumoylation.Cell Cycle. 2013 Nov 1;12(21):3442-7. doi: 10.4161/cc.26465. Epub 2013 Sep 18. Cell Cycle. 2013. PMID: 24047694 Free PMC article.
-
Homologous recombination and its regulation.Nucleic Acids Res. 2012 Jul;40(13):5795-818. doi: 10.1093/nar/gks270. Epub 2012 Mar 30. Nucleic Acids Res. 2012. PMID: 22467216 Free PMC article. Review.
-
Sumoylation of eIF4A2 affects stress granule formation.J Cell Sci. 2016 Jun 15;129(12):2407-15. doi: 10.1242/jcs.184614. Epub 2016 May 9. J Cell Sci. 2016. PMID: 27160682 Free PMC article.
References
-
- Taylor E.M. and Lehmann,A.R. (1998) Conservation of eukaryotic DNA repair mechanisms. Int. J. Radiat. Biol., 74, 277–286. - PubMed
-
- Caspari T. and Carr,A.M. (1999) DNA structure checkpoint pathways in Schizosaccharomyces pombe. Biochimie, 81, 173–181. - PubMed
-
- al-Khodairy F., Enoch,T., Hagan,I.M. and Carr,A.M. (1995) The Schizosaccharomyces pombe hus5 gene encodes a ubiquitin conjugating enzyme required for normal mitosis. J. Cell Sci., 108, 475–486. - PubMed
-
- Johnson E.S. and Blobel,G. (1997) Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem., 272, 26799–26802. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

