Box C/D small nucleolar RNA trafficking involves small nucleolar RNP proteins, nucleolar factors and a novel nuclear domain
- PMID: 11574480
- PMCID: PMC125276
- DOI: 10.1093/emboj/20.19.5480
Box C/D small nucleolar RNA trafficking involves small nucleolar RNP proteins, nucleolar factors and a novel nuclear domain
Abstract
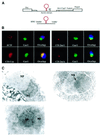
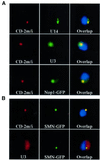
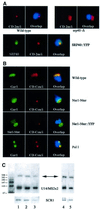
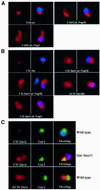
Nucleolar localization of box C/D small nucleolar (sno) RNAs requires the box C/D motif and, in vertebrates, involves transit through Cajal bodies (CB). We report that in yeast, overexpression of a box C/D reporter leads to a block in the localization pathway with snoRNA accumulation in a specific sub-nucleolar structure, the nucleolar body (NB). The human survival of motor neuron protein (SMN), a marker of gems/CB, specifically localizes to the NB when expressed in yeast, supporting similarities between these structures. Box C/D snoRNA accumulation in the NB was decreased by mutation of Srp40 and increased by mutation of Nsr1p, two related nucleolar proteins that are homologous to human Nopp140 and nucleolin, respectively. Box C/D snoRNAs also failed to accumulate in the NB, and became delocalized to the nucleoplasm, upon depletion of any of the core snoRNP proteins, Nop1p/fibrillarin, Snu13p, Nop56p and Nop5p/Nop58p. We conclude that snoRNP assembly occurs either in the nucleoplasm, or during transit of snoRNAs through the NB, followed by routing of the complete snoRNP to functional sites of ribosome synthesis.
Figures

Similar articles
-
[Small nucleolar RNAs].Mol Biol (Mosk). 2007 Mar-Apr;41(2):246-59. Mol Biol (Mosk). 2007. PMID: 17514894 Review. Russian.
-
Implication of the box C/D snoRNP assembly factor Rsa1p in U3 snoRNP assembly.Nucleic Acids Res. 2017 Jul 7;45(12):7455-7473. doi: 10.1093/nar/gkx424. Nucleic Acids Res. 2017. PMID: 28505348 Free PMC article.
-
Synthesis and assembly of the box C+D small nucleolar RNPs.Mol Cell Biol. 2000 Apr;20(8):2650-9. doi: 10.1128/MCB.20.8.2650-2659.2000. Mol Cell Biol. 2000. PMID: 10733567 Free PMC article.
-
A well-connected and conserved nucleoplasmic helicase is required for production of box C/D and H/ACA snoRNAs and localization of snoRNP proteins.Mol Cell Biol. 2001 Nov;21(22):7731-46. doi: 10.1128/MCB.21.22.7731-7746.2001. Mol Cell Biol. 2001. PMID: 11604509 Free PMC article.
-
Nucleolar RNPs: from genes to functional snoRNAs in plants.Biochem Soc Trans. 2010 Apr;38(2):672-6. doi: 10.1042/BST0380672. Biochem Soc Trans. 2010. PMID: 20298241 Review.
Cited by
-
RNA modification in Cajal bodies.RNA Biol. 2017 Jun 3;14(6):693-700. doi: 10.1080/15476286.2016.1249091. Epub 2016 Oct 24. RNA Biol. 2017. PMID: 27775477 Free PMC article. Review.
-
CRM1 plays a nuclear role in transporting snoRNPs to nucleoli in higher eukaryotes.Nucleus. 2012 Mar 1;3(2):132-7. doi: 10.4161/nucl.19266. Epub 2012 Mar 1. Nucleus. 2012. PMID: 22555597 Free PMC article. Review.
-
Mammalian and yeast U3 snoRNPs are matured in specific and related nuclear compartments.EMBO J. 2002 Jun 3;21(11):2736-45. doi: 10.1093/emboj/21.11.2736. EMBO J. 2002. PMID: 12032086 Free PMC article.
-
Conventional and nonconventional roles of the nucleolus.Int Rev Cytol. 2002;219:199-266. doi: 10.1016/s0074-7696(02)19014-0. Int Rev Cytol. 2002. PMID: 12211630 Free PMC article. Review.
-
Liquid-Liquid Phase Separation: Unraveling the Enigma of Biomolecular Condensates in Microbial Cells.Front Microbiol. 2021 Oct 25;12:751880. doi: 10.3389/fmicb.2021.751880. eCollection 2021. Front Microbiol. 2021. PMID: 34759902 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

