The tyrosyl-DNA phosphodiesterase Tdp1 is a member of the phospholipase D superfamily
- PMID: 11572945
- PMCID: PMC59758
- DOI: 10.1073/pnas.211429198
The tyrosyl-DNA phosphodiesterase Tdp1 is a member of the phospholipase D superfamily
Abstract
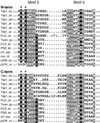
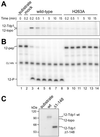
The phospholipase D (PLD) superfamily is a diverse group of proteins that includes enzymes involved in phospholipid metabolism, a bacterial toxin, poxvirus envelope proteins, and bacterial nucleases. Based on sequence comparisons, we show here that the tyrosyl-DNA phosphodiesterase (Tdp1) that has been implicated in the repair of topoisomerase I covalent complexes with DNA contains two unusual HKD signature motifs that place the enzyme in a distinct class within the PLD superfamily. Mutagenesis studies with the human enzyme in which the invariant histidines and lysines of the HKD motifs are changed confirm that these highly conserved residues are essential for Tdp1 activity. Furthermore, we show that, like other members of the family for which it has been examined, the reaction involves the formation of an intermediate in which the cleaved substrate is covalently linked to the enzyme. These results reveal that the hydrolytic reaction catalyzed by Tdp1 occurs by the phosphoryl transfer chemistry that is common to all members of the PLD superfamily.
Figures


Similar articles
-
The crystal structure of human tyrosyl-DNA phosphodiesterase, Tdp1.Structure. 2002 Feb;10(2):237-48. doi: 10.1016/s0969-2126(02)00707-4. Structure. 2002. PMID: 11839309
-
Structure-function studies of a plant tyrosyl-DNA phosphodiesterase provide novel insights into DNA repair mechanisms of Arabidopsis thaliana.Biochem J. 2012 Apr 1;443(1):49-56. doi: 10.1042/BJ20111308. Biochem J. 2012. PMID: 22214184 Free PMC article.
-
Analysis of the active-site mechanism of tyrosyl-DNA phosphodiesterase I: a member of the phospholipase D superfamily.J Mol Biol. 2012 Jan 27;415(4):741-58. doi: 10.1016/j.jmb.2011.11.044. Epub 2011 Dec 6. J Mol Biol. 2012. PMID: 22155078 Free PMC article.
-
Tyrosyl-DNA phosphodiesterase I resolves both naturally and chemically induced DNA adducts and its potential as a therapeutic target.Drug Metab Rev. 2014 Nov;46(4):494-507. doi: 10.3109/03602532.2014.971957. Epub 2014 Oct 20. Drug Metab Rev. 2014. PMID: 25327705 Review.
-
Tyrosyl-DNA Phosphodiesterase I N-Terminal Domain Modifications and Interactions Regulate Cellular Function.Genes (Basel). 2019 Nov 6;10(11):897. doi: 10.3390/genes10110897. Genes (Basel). 2019. PMID: 31698852 Free PMC article. Review.
Cited by
-
Tyrosyl-DNA phosphodiesterase 1 initiates repair of apurinic/apyrimidinic sites.Biochimie. 2012 Aug;94(8):1749-53. doi: 10.1016/j.biochi.2012.04.004. Epub 2012 Apr 12. Biochimie. 2012. PMID: 22522093 Free PMC article.
-
Human Tdp1 cleaves a broad spectrum of substrates, including phosphoamide linkages.J Biol Chem. 2005 Oct 28;280(43):36518-28. doi: 10.1074/jbc.M508898200. Epub 2005 Aug 31. J Biol Chem. 2005. PMID: 16141202 Free PMC article.
-
SCAN1 mutant Tdp1 accumulates the enzyme--DNA intermediate and causes camptothecin hypersensitivity.EMBO J. 2005 Jun 15;24(12):2224-33. doi: 10.1038/sj.emboj.7600694. Epub 2005 May 26. EMBO J. 2005. PMID: 15920477 Free PMC article.
-
N-terminal domain of tyrosyl-DNA phosphodiesterase I regulates topoisomerase I-induced toxicity in cells.Sci Rep. 2023 Jan 25;13(1):1377. doi: 10.1038/s41598-023-28564-6. Sci Rep. 2023. PMID: 36697463 Free PMC article.
-
DNA-protein cross-link repair: what do we know now?Cell Biosci. 2020 Jan 7;10:3. doi: 10.1186/s13578-019-0366-z. eCollection 2020. Cell Biosci. 2020. PMID: 31921408 Free PMC article. Review.
References
-
- Morris A J, Engebrecht J, Frohman M A. Trends Pharmacol Sci. 1996;17:182–185. - PubMed
-
- Koonin E V. Trends Biochem Sci. 1996;21:242–243. - PubMed
-
- Stuckey J A, Dixon J E. Nat Struct Biol. 1999;6:278–284. - PubMed
-
- Leiros I, Secundo F, Zambonelli C, Servi S, Hough E. Struct Fold Des. 2000;8:655–667. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

