An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells
- PMID: 11564755
- PMCID: PMC2150808
- DOI: 10.1083/jcb.200101081
An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells
Abstract
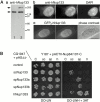
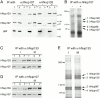
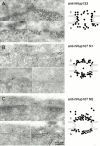
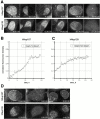
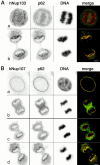
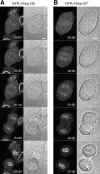
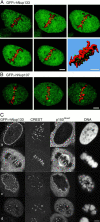
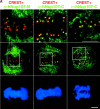
The nuclear pore complexes (NPCs) are evolutionarily conserved assemblies that allow traffic between the cytoplasm and the nucleus. In this study, we have identified and characterized a novel human nuclear pore protein, hNup133, through its homology with the Saccharomyces cerevisiae nucleoporin scNup133. Two-hybrid screens and immunoprecipitation experiments revealed a direct and evolutionarily conserved interaction between Nup133 and Nup84/Nup107 and indicated that hNup133 and hNup107 are part of a NPC subcomplex that contains two other nucleoporins (the previously characterized hNup96 and a novel nucleoporin designated as hNup120) homologous to constituents of the scNup84 subcomplex. We further demonstrate that hNup133 and hNup107 are localized on both sides of the NPC to which they are stably associated at interphase, remain associated as part of a NPC subcomplex during mitosis, and are targeted at early stages to the reforming nuclear envelope. Throughout mitosis, a fraction of hNup133 and hNup107 localizes to the kinetochores, thus revealing an unexpected connection between structural NPCs constituents and kinetochores. Photobleaching experiments further showed that the mitotic cytoplasm contains kinetochore-binding competent hNup133 molecules and that in contrast to its stable association with the NPCs the interaction of this nucleoporin with kinetochores is dynamic.
Figures


Similar articles
-
Nucleoporins: leaving the nuclear pore complex for a successful mitosis.Cell Signal. 2011 Oct;23(10):1555-62. doi: 10.1016/j.cellsig.2011.05.023. Epub 2011 Jun 13. Cell Signal. 2011. PMID: 21683138 Review.
-
ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division.Proc Natl Acad Sci U S A. 2006 Nov 21;103(47):17801-6. doi: 10.1073/pnas.0608484103. Epub 2006 Nov 10. Proc Natl Acad Sci U S A. 2006. PMID: 17098863 Free PMC article.
-
Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex.Proc Natl Acad Sci U S A. 2003 Feb 4;100(3):981-5. doi: 10.1073/pnas.252749899. Epub 2003 Jan 27. Proc Natl Acad Sci U S A. 2003. PMID: 12552102 Free PMC article.
-
Dbp5, a DEAD-box protein required for mRNA export, is recruited to the cytoplasmic fibrils of nuclear pore complex via a conserved interaction with CAN/Nup159p.EMBO J. 1999 Aug 2;18(15):4332-47. doi: 10.1093/emboj/18.15.4332. EMBO J. 1999. PMID: 10428971 Free PMC article.
-
High resolution analysis of mammalian nuclear structure throughout the cell cycle: implications for nuclear pore complex assembly during interphase and mitosis.Can J Physiol Pharmacol. 2006 Mar-Apr;84(3-4):423-30. doi: 10.1139/y05-148. Can J Physiol Pharmacol. 2006. PMID: 16902587 Review.
Cited by
-
Sm protein down-regulation leads to defects in nuclear pore complex disassembly and distribution in C. elegans embryos.Dev Biol. 2012 May 15;365(2):445-57. doi: 10.1016/j.ydbio.2012.02.036. Epub 2012 Mar 8. Dev Biol. 2012. PMID: 22426005 Free PMC article.
-
Direct interaction with nup153 mediates binding of Tpr to the periphery of the nuclear pore complex.Mol Biol Cell. 2003 May;14(5):1923-40. doi: 10.1091/mbc.e02-09-0620. Mol Biol Cell. 2003. PMID: 12802065 Free PMC article.
-
Nucleoporins as components of the nuclear pore complex core structure and Tpr as the architectural element of the nuclear basket.Mol Biol Cell. 2004 Sep;15(9):4261-77. doi: 10.1091/mbc.e04-03-0165. Epub 2004 Jun 30. Mol Biol Cell. 2004. PMID: 15229283 Free PMC article.
-
Capture of AT-rich chromatin by ELYS recruits POM121 and NDC1 to initiate nuclear pore assembly.Mol Biol Cell. 2008 Sep;19(9):3982-96. doi: 10.1091/mbc.e08-01-0012. Epub 2008 Jul 2. Mol Biol Cell. 2008. PMID: 18596237 Free PMC article.
-
Functional study of a novel missense single-nucleotide variant of NUP107 in two daughters of Mexican origin with premature ovarian insufficiency.Mol Genet Genomic Med. 2018 Mar;6(2):276-281. doi: 10.1002/mgg3.345. Epub 2018 Jan 24. Mol Genet Genomic Med. 2018. PMID: 29363275 Free PMC article.
References
-
- Adams, R.R., M. Carmena, and W.C. Earnshaw. 2001. Chromosomal passengers and the (aurora) ABCs of mitosis. Trends Cell Biol. 11:49–54. - PubMed
-
- Azuma, Y., and M. Dasso. 2000. The role of Ran in nuclear function. Curr. Opin. Cell Biol. 12:302–330. - PubMed
-
- Belgareh, N., and V. Doye. 1999. Yeast and vertebrate nuclear-pore complexes: evolutionary conserved, yet divergent macromolecular assemblies. Protoplasma. 209:133–143.
-
- Bodoor, K., S. Shaikh, D. Salina, W.H. Raharjo, R. Bastos, M. Lohka, and B. Burke. 1999. Sequential recruitment of NPC proteins to the nuclear periphery at the end of mitosis. J. Cell Sci. 112:2253–2264. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

