In vivo transposition of Minos, a Drosophila mobile element, in mammalian tissues
- PMID: 11562481
- PMCID: PMC58754
- DOI: 10.1073/pnas.201392398
In vivo transposition of Minos, a Drosophila mobile element, in mammalian tissues
Abstract
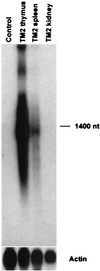
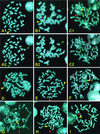
Transposable elements have been used widely in the past 20 years for gene transfer and insertional mutagenesis in Drosophila. Transposon-based technology for gene manipulation and genomic analysis currently is being adopted for vertebrates. We tested the ability of Minos, a DNA transposon from Drosophila hydei, to transpose in mouse tissues. Two transgenic mouse lines were crossed, one expressing Minos transposase in lymphocytes under the control of the CD2 promoter/locus control region and another carrying a nonautonomous Minos transposon. Only mice containing both transgenes show excision of the transposon and transposition into new chromosomal sites in thymus and spleen cells. In addition, expression of Minos transposase in embryonic fibroblast cell lines derived from a transposon-carrying transgenic mouse resulted in excision of the transposon. These results are a first step toward a reversible insertional mutagenesis system in the mouse, opening the way to develop powerful technologies for functional genomic analysis in mammals.
Figures

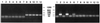

Similar articles
-
Transposition of the Drosophila hydei Minos transposon in the mouse germ line.Genomics. 2003 Feb;81(2):108-11. doi: 10.1016/s0888-7543(02)00030-7. Genomics. 2003. PMID: 12620388
-
Efficient transposition of a single Minos transposon copy in the genome of the ascidian Ciona intestinalis with a transgenic line expressing transposase in eggs.Dev Dyn. 2010 Apr;239(4):1076-88. doi: 10.1002/dvdy.22254. Dev Dyn. 2010. PMID: 20186916
-
Mobilization of a Minos transposon in Drosophila melanogaster chromosomes and chromatid repair by heteroduplex formation.Genetics. 1997 Feb;145(2):267-79. doi: 10.1093/genetics/145.2.267. Genetics. 1997. PMID: 9071583 Free PMC article.
-
Germline transgenesis and insertional mutagenesis in the ascidian Ciona intestinalis.Dev Dyn. 2007 Jul;236(7):1758-67. doi: 10.1002/dvdy.21111. Dev Dyn. 2007. PMID: 17342755 Review.
-
Sleeping Beauty Transposition.Microbiol Spectr. 2015 Apr;3(2):MDNA3-0042-2014. doi: 10.1128/microbiolspec.MDNA3-0042-2014. Microbiol Spectr. 2015. PMID: 26104705 Review.
Cited by
-
Cancer gene discovery in mouse and man.Biochim Biophys Acta. 2009 Dec;1796(2):140-61. doi: 10.1016/j.bbcan.2009.03.001. Epub 2009 Mar 12. Biochim Biophys Acta. 2009. PMID: 19285540 Free PMC article. Review.
-
Nonhomologous-end-joining factors regulate DNA repair fidelity during Sleeping Beauty element transposition in mammalian cells.Mol Cell Biol. 2003 Dec;23(23):8505-18. doi: 10.1128/MCB.23.23.8505-8518.2003. Mol Cell Biol. 2003. PMID: 14612396 Free PMC article.
-
Efficient transformation of the beetle Tribolium castaneum using the Minos transposable element: quantitative and qualitative analysis of genomic integration events.Genetics. 2004 Jun;167(2):737-46. doi: 10.1534/genetics.103.023085. Genetics. 2004. PMID: 15238525 Free PMC article.
-
The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes.Genetics. 2004 Jun;167(2):761-81. doi: 10.1534/genetics.104.026427. Genetics. 2004. PMID: 15238527 Free PMC article.
-
PBmice: an integrated database system of piggyBac (PB) insertional mutations and their characterizations in mice.Nucleic Acids Res. 2008 Jan;36(Database issue):D729-34. doi: 10.1093/nar/gkm790. Epub 2007 Oct 11. Nucleic Acids Res. 2008. PMID: 17932058 Free PMC article.
References
-
- Spradling A C, Rubin G M. Science. 1982;218:341–347. - PubMed
-
- Walbot V. Curr Opin Plant Biol. 2000;3:103–107. - PubMed
-
- Raz E, van Luenen H G, Schaerringer B, Plasterk R H A, Driever W. Curr Biol. 1998;8:82–88. - PubMed
-
- Sherman A, Dawson A, Mather C, Gilhooley H, Li Y, Mitchell R, Finnegan D, Sang H. Nat Biotechnol. 1998;16:1050–1053. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

