A moving DNA replication factory in Caulobacter crescentus
- PMID: 11532959
- PMCID: PMC125615
- DOI: 10.1093/emboj/20.17.4952
A moving DNA replication factory in Caulobacter crescentus
Abstract
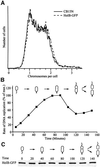
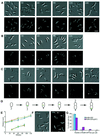
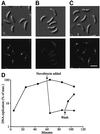
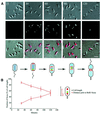
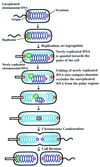
The in vivo intracellular location of components of the Caulobacter replication apparatus was visualized during the cell cycle. Replisome assembly occurs at the chromosomal origin located at the stalked cell pole, coincident with the initiation of DNA replication. The replisome gradually moves to midcell as DNA replication proceeds and disassembles upon completion of DNA replication. Although the newly replicated origin regions of the chromosome are rapidly moved to opposite cell poles by an active process, the replisome appears to be an untethered replication factory that is passively displaced towards the center of the cell by the newly replicated DNA. These results are consistent with a model in which unreplicated DNA is pulled into the replication factory and newly replicated DNA is bidirectionally extruded from the complex, perhaps contributing to chromosome segregation.
Figures


Similar articles
-
Caulobacter requires a dedicated mechanism to initiate chromosome segregation.Proc Natl Acad Sci U S A. 2008 Oct 7;105(40):15435-40. doi: 10.1073/pnas.0807448105. Epub 2008 Sep 29. Proc Natl Acad Sci U S A. 2008. PMID: 18824683 Free PMC article.
-
A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole.Cell. 2008 Sep 19;134(6):945-55. doi: 10.1016/j.cell.2008.07.015. Cell. 2008. PMID: 18805088 Free PMC article.
-
Coordination between chromosome replication, segregation, and cell division in Caulobacter crescentus.J Bacteriol. 2006 Mar;188(6):2244-53. doi: 10.1128/JB.188.6.2244-2253.2006. J Bacteriol. 2006. PMID: 16513754 Free PMC article.
-
Cell cycle regulation in Caulobacter: location, location, location.J Cell Sci. 2007 Oct 15;120(Pt 20):3501-7. doi: 10.1242/jcs.005967. J Cell Sci. 2007. PMID: 17928306 Review.
-
Regulation of chromosomal replication in Caulobacter crescentus.Plasmid. 2012 Mar;67(2):76-87. doi: 10.1016/j.plasmid.2011.12.007. Epub 2011 Dec 29. Plasmid. 2012. PMID: 22227374 Review.
Cited by
-
Spatiotemporal control of PopZ localization through cell cycle-coupled multimerization.J Cell Biol. 2013 Jun 10;201(6):827-41. doi: 10.1083/jcb.201303036. J Cell Biol. 2013. PMID: 23751494 Free PMC article.
-
Where and When Bacterial Chromosome Replication Starts: A Single Cell Perspective.Front Microbiol. 2018 Nov 26;9:2819. doi: 10.3389/fmicb.2018.02819. eCollection 2018. Front Microbiol. 2018. PMID: 30534115 Free PMC article. Review.
-
Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication.Proc Natl Acad Sci U S A. 2004 Jun 22;101(25):9257-62. doi: 10.1073/pnas.0402606101. Epub 2004 Jun 3. Proc Natl Acad Sci U S A. 2004. PMID: 15178755 Free PMC article.
-
The topoisomerase IV ParC subunit colocalizes with the Caulobacter replisome and is required for polar localization of replication origins.Proc Natl Acad Sci U S A. 2004 Jun 22;101(25):9251-6. doi: 10.1073/pnas.0402567101. Epub 2004 Jun 3. Proc Natl Acad Sci U S A. 2004. PMID: 15178756 Free PMC article.
-
Organization of human replicon: singles or zipping couples?J Struct Biol. 2009 Mar;165(3):204-13. doi: 10.1016/j.jsb.2008.11.004. Epub 2008 Dec 3. J Struct Biol. 2009. PMID: 19063972 Free PMC article.
References
-
- Cook P.R. (1999) The organization of replication and transcription. Science, 284, 1790–1795. - PubMed
-
- Davey M.J. and O’Donnell,M. (2000) Mechanisms of DNA replication. Curr. Opin. Chem. Biol., 4, 581–586. - PubMed
-
- Dingman C.W. (1974) Bidirectional chromosome replication: some topological considerations. J. Theor. Biol., 43, 187–195. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

