Epstein-Barr nuclear antigen 1 binds and destabilizes nucleosomes at the viral origin of latent DNA replication
- PMID: 11522821
- PMCID: PMC55891
- DOI: 10.1093/nar/29.17.3520
Epstein-Barr nuclear antigen 1 binds and destabilizes nucleosomes at the viral origin of latent DNA replication
Abstract
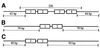
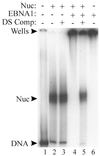
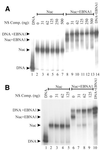
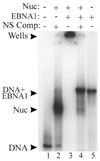
The EBNA1 protein of Epstein-Barr virus (EBV) activates latent-phase DNA replication by an unknown mechanism that involves binding to four recognition sites in the dyad symmetry (DS) element of the viral latent origin of DNA replication. Since EBV episomes are assembled into nucleosomes, we have examined the ability of Epstein-Barr virus nuclear antigen 1 (EBNA1) to interact with the DS element when it is assembled into a nucleosome core particle. EBNA1 bound to its recognition sites within this nucleosome, forming a ternary complex, and displaced the histone octamer upon competitor DNA challenge. The DNA binding and dimerization region of EBNA1 was sufficient for nucleosome binding and destabilization. Although EBNA1 was able to bind to nucleosomes containing two recognition sites from the DS element positioned at the edge of the nucleosome, nucleosome destabilization was only observed when all four sites of the DS element were present. Our results indicate that the presence of a nucleosome at the viral origin will not prevent EBNA1 binding to its recognition sites. In addition, since four EBNA1 recognition sites are required for both nucleosome destabilization and efficient origin activation, our findings also suggest that nucleosome destabilization by EBNA1 is important for origin activation.
Figures



Similar articles
-
Structural Basis for Cooperative Binding of EBNA1 to the Epstein-Barr Virus Dyad Symmetry Minimal Origin of Replication.J Virol. 2019 Sep 30;93(20):e00487-19. doi: 10.1128/JVI.00487-19. Print 2019 Oct 15. J Virol. 2019. PMID: 31142669 Free PMC article.
-
Cryo-EM Structure and Functional Studies of EBNA1 Binding to the Family of Repeats and Dyad Symmetry Elements of Epstein-Barr Virus oriP.J Virol. 2022 Sep 14;96(17):e0094922. doi: 10.1128/jvi.00949-22. Epub 2022 Aug 29. J Virol. 2022. PMID: 36037477 Free PMC article.
-
The 2.2 A structure of a permanganate-sensitive DNA site bound by the Epstein-Barr virus origin binding protein, EBNA1.J Mol Biol. 1998 Dec 18;284(5):1273-8. doi: 10.1006/jmbi.1998.2247. J Mol Biol. 1998. PMID: 9878348 Review.
-
Stabilization of the EBNA1 protein on the Epstein-Barr virus latent origin of DNA replication by a DNA looping mechanism.J Biol Chem. 1994 Jan 14;269(2):1057-62. J Biol Chem. 1994. PMID: 8288561
-
Replication licensing of the EBV oriP minichromosome.Curr Top Microbiol Immunol. 2001;258:13-33. doi: 10.1007/978-3-642-56515-1_2. Curr Top Microbiol Immunol. 2001. PMID: 11443858 Review.
Cited by
-
Chromatin organization of gammaherpesvirus latent genomes.Biochim Biophys Acta. 2010 Mar-Apr;1799(3-4):236-45. doi: 10.1016/j.bbagrm.2009.10.004. Epub 2009 Oct 22. Biochim Biophys Acta. 2010. PMID: 19853673 Free PMC article. Review.
-
The Epstein-Barr Virus EBNA1 Protein.Scientifica (Cairo). 2012;2012:438204. doi: 10.6064/2012/438204. Epub 2012 Dec 19. Scientifica (Cairo). 2012. PMID: 24278697 Free PMC article. Review.
-
Mitotic chromosome interactions of Epstein-Barr nuclear antigen 1 (EBNA1) and human EBNA1-binding protein 2 (EBP2).J Cell Sci. 2009 Dec 1;122(Pt 23):4341-50. doi: 10.1242/jcs.060913. Epub 2009 Nov 3. J Cell Sci. 2009. PMID: 19887584 Free PMC article.
-
Epigenetic regulation of EBV persistence and oncogenesis.Semin Cancer Biol. 2014 Jun;26:22-9. doi: 10.1016/j.semcancer.2014.01.003. Epub 2014 Jan 24. Semin Cancer Biol. 2014. PMID: 24468737 Free PMC article. Review.
-
Similarities between the Epstein-Barr Virus (EBV) Nuclear Protein EBNA1 and the Pioneer Transcription Factor FoxA: Is EBNA1 a "Bookmarking" Oncoprotein that Alters the Host Cell Epigenotype?Pathogens. 2012 Sep 17;1(1):37-51. doi: 10.3390/pathogens1010037. Pathogens. 2012. PMID: 25436603 Free PMC article. Review.
References
-
- Kieff E. (1996) Epstein–Barr virus and its replication. In Fields,B.N., Knipe,D.M. and Howley,P.M. (eds), Fields Virology, 3rd edition. Lippincott-Raven Publishers, PA, pp. 2343–2396.
-
- Yates J.L. (1996) Epstein–Barr virus DNA replication. In DePamphilis,M.L. (ed.), DNA Replication in Eukaryotic Cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 751–773.
-
- Rawlins D.R., Milman,G., Hayward,S.D. and Hayward,G.S. (1985) Sequence-specific DNA binding of the Epstein-Barr virus nuclear antigen (EBNA1) to clustered sites in the plasmid maintenance region. Cell, 42, 859–868. - PubMed

