Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation
- PMID: 11511539
- PMCID: PMC312758
- DOI: 10.1101/gad.906601
Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation
Abstract
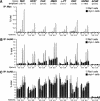
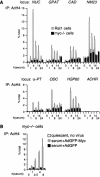
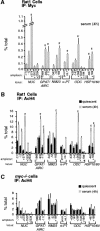
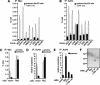
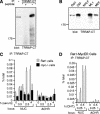
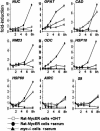
The Myc protein binds DNA and activates transcription by mechanisms that are still unclear. We used chromatin immunoprecipitation (ChIP) to evaluate Myc-dependent changes in histone acetylation at seven target loci. Upon serum stimulation of Rat1 fibroblasts, Myc associated with chromatin, histone H4 became locally hyperacetylated, and gene expression was induced. These responses were lost or severely impaired in Myc-deficient cells, but were restored by adenoviral delivery of Myc simultaneous with mitogenic stimulation. When targeted to chromatin in the absence of mitogens, Myc directly induced H4 acetylation. In addition, Myc recruited TRRAP to chromatin, consistent with a role for this cofactor in histone acetylation. Finally, unlike serum, Myc alone was very inefficient in inducing expression of most target genes. Myc therefore governs a step, most likely H4 acetylation, that is required but not sufficient for transcriptional activation. We propose that Myc acts as a permissive factor, allowing additional signals to activate target promoters.
Figures



Similar articles
-
Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter.Genes Dev. 2001 Aug 15;15(16):2042-7. doi: 10.1101/gad.907901. Genes Dev. 2001. PMID: 11511535 Free PMC article.
-
The mouse C/EBPdelta gene promoter is regulated by STAT3 and Sp1 transcriptional activators, chromatin remodeling and c-Myc repression.J Cell Biochem. 2007 Dec 1;102(5):1256-70. doi: 10.1002/jcb.21356. J Cell Biochem. 2007. PMID: 17471507
-
Myc-binding-site recognition in the human genome is determined by chromatin context.Nat Cell Biol. 2006 Jul;8(7):764-70. doi: 10.1038/ncb1434. Epub 2006 Jun 11. Nat Cell Biol. 2006. PMID: 16767079
-
Deconstructing myc.Genes Dev. 2001 Aug 15;15(16):2023-30. doi: 10.1101/gad928101. Genes Dev. 2001. PMID: 11511533 Review. No abstract available.
-
Transcriptional activation by the Myc oncoprotein.Curr Top Microbiol Immunol. 2006;302:33-50. doi: 10.1007/3-540-32952-8_2. Curr Top Microbiol Immunol. 2006. PMID: 16620024 Review.
Cited by
-
Sirt1 ablation promotes stress-induced loss of epigenetic and genomic hematopoietic stem and progenitor cell maintenance.J Exp Med. 2013 May 6;210(5):987-1001. doi: 10.1084/jem.20121608. Epub 2013 Apr 29. J Exp Med. 2013. PMID: 23630229 Free PMC article.
-
MacroH2A1 regulates the balance between self-renewal and differentiation commitment in embryonic and adult stem cells.Mol Cell Biol. 2012 Apr;32(8):1442-52. doi: 10.1128/MCB.06323-11. Epub 2012 Feb 13. Mol Cell Biol. 2012. PMID: 22331466 Free PMC article.
-
A human B-cell interactome identifies MYB and FOXM1 as master regulators of proliferation in germinal centers.Mol Syst Biol. 2010 Jun 8;6:377. doi: 10.1038/msb.2010.31. Mol Syst Biol. 2010. PMID: 20531406 Free PMC article.
-
An early Myc-dependent transcriptional program orchestrates cell growth during B-cell activation.EMBO Rep. 2019 Sep;20(9):e47987. doi: 10.15252/embr.201947987. Epub 2019 Jul 23. EMBO Rep. 2019. PMID: 31334602 Free PMC article.
-
Pendrin is a novel in vivo downstream target gene of the TTF-1/Nkx-2.1 homeodomain transcription factor in differentiated thyroid cells.Mol Cell Biol. 2005 Nov;25(22):10171-82. doi: 10.1128/MCB.25.22.10171-10182.2005. Mol Cell Biol. 2005. PMID: 16260629 Free PMC article.
References
-
- Agalioti T, Lomvardas S, Parekh B, Yie J, Maniatis T, Thanos D. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell. 2000;103:667–678. - PubMed
-
- Amati B, Frank SR, Donjerkovic D, Taubert S. Function of the c-Myc oncoprotein in chromatin remodeling and transcription. Biochim Biophys Acta. 2001;1471:M135–M145. - PubMed
-
- Aparicio OM. Characterization of proteins bound to chromatin by immunoprecipitation from whole-cell extracts. In: Ausubel FM, Brent R, Kingston RE, Moore DM, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. New York: John Wiley; 1999. pp. 21.23.21–21.23.12.
-
- Ayer DE, Eisenman RN. A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation. Genes & Dev. 1993;7:2110–2119. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
