RNA-mediated interaction of Cajal bodies and U2 snRNA genes
- PMID: 11489914
- PMCID: PMC2196410
- DOI: 10.1083/jcb.200105084
RNA-mediated interaction of Cajal bodies and U2 snRNA genes
Abstract
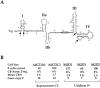
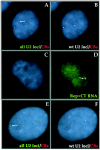
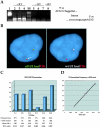
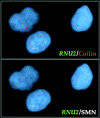
Cajal bodies (CBs) are nuclear structures involved in RNA metabolism that accumulate high concentrations of small nuclear ribonucleoproteins (snRNPs). Notably, CBs preferentially associate with specific genomic loci in interphase human cells, including several snRNA and histone gene clusters. To uncover functional elements involved in the interaction of genes and CBs, we analyzed the expression and subcellular localization of stably transfected artificial arrays of U2 snRNA genes. Although promoter substitution arrays colocalized with CBs, constructs containing intragenic deletions did not. Additional experiments identified factors within CBs that are important for association with the native U2 genes. Inhibition of nuclear export or targeted degradation of U2 snRNPs caused a marked decrease in the levels of U2 snRNA in CBs and strongly disrupted the interaction with U2 genes. Together, the results illustrate a specific requirement for both the snRNA transcripts as well as the presence of snRNPs (or snRNP proteins) within CBs. Our data thus provide significant insight into the mechanism of CB interaction with snRNA loci, strengthening the putative role for this nuclear suborganelle in snRNP biogenesis.
Figures


Similar articles
-
The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze.Chromosoma. 2006 Oct;115(5):343-54. doi: 10.1007/s00412-006-0056-6. Epub 2006 Mar 31. Chromosoma. 2006. PMID: 16575476 Review.
-
Interactions of U2 gene loci and their nuclear transcripts with Cajal (coiled) bodies: evidence for PreU2 within Cajal bodies.Mol Biol Cell. 2000 Sep;11(9):2987-98. doi: 10.1091/mbc.11.9.2987. Mol Biol Cell. 2000. PMID: 10982395 Free PMC article.
-
A role for Cajal bodies in the final steps of U2 snRNP biogenesis.J Cell Sci. 2004 Sep 1;117(Pt 19):4423-33. doi: 10.1242/jcs.01308. Epub 2004 Aug 17. J Cell Sci. 2004. PMID: 15316075
-
Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins.Proc Natl Acad Sci U S A. 2005 Nov 29;102(48):17372-7. doi: 10.1073/pnas.0508947102. Epub 2005 Nov 21. Proc Natl Acad Sci U S A. 2005. PMID: 16301532 Free PMC article.
-
The SMN complex: an assembly machine for RNPs.Cold Spring Harb Symp Quant Biol. 2006;71:313-20. doi: 10.1101/sqb.2006.71.001. Cold Spring Harb Symp Quant Biol. 2006. PMID: 17381311 Review.
Cited by
-
The Cajal Body Protein WRAP53β Prepares the Scene for Repair of DNA Double-Strand Breaks by Regulating Local Ubiquitination.Front Mol Biosci. 2019 Jul 4;6:51. doi: 10.3389/fmolb.2019.00051. eCollection 2019. Front Mol Biosci. 2019. PMID: 31334247 Free PMC article. Review.
-
The Cajal body: a meeting place for spliceosomal snRNPs in the nuclear maze.Chromosoma. 2006 Oct;115(5):343-54. doi: 10.1007/s00412-006-0056-6. Epub 2006 Mar 31. Chromosoma. 2006. PMID: 16575476 Review.
-
Functional nuclear retention of pre-mRNA involving Cajal bodies during meiotic prophase in European larch (Larix decidua).Plant Cell. 2022 May 24;34(6):2404-2423. doi: 10.1093/plcell/koac091. Plant Cell. 2022. PMID: 35294035 Free PMC article.
-
Cajal body surveillance of U snRNA export complex assembly.J Cell Biol. 2010 Aug 23;190(4):603-12. doi: 10.1083/jcb.201004109. J Cell Biol. 2010. PMID: 20733056 Free PMC article.
-
High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli.Mol Biol Cell. 2010 Nov 1;21(21):3735-48. doi: 10.1091/mbc.E10-06-0508. Epub 2010 Sep 8. Mol Biol Cell. 2010. PMID: 20826608 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

