Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential
- PMID: 11483586
- PMCID: PMC311097
- DOI: 10.1101/gr.184001
Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential
Abstract
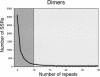
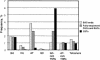
A total of 57.8 Mb of publicly available rice (Oryza sativa L.) DNA sequence was searched to determine the frequency and distribution of different simple sequence repeats (SSRs) in the genome. SSR loci were categorized into two groups based on the length of the repeat motif. Class I, or hypervariable markers, consisted of SSRs > or =20 bp, and Class II, or potentially variable markers, consisted of SSRs > or =12 bp <20 bp. The occurrence of Class I SSRs in end-sequences of EcoRI- and HindIII-digested BAC clones was one SSR per 40 Kb, whereas in continuous genomic sequence (represented by 27 fully sequenced BAC and PAC clones), the frequency was one SSR every 16 kb. Class II SSRs were estimated to occur every 3.7 kb in BAC ends and every 1.9 kb in fully sequenced BAC and PAC clones. GC-rich trinucleotide repeats (TNRs) were most abundant in protein-coding portions of ESTs and in fully sequenced BACs and PACs, whereas AT-rich TNRs showed no such preference, and di- and tetranucleotide repeats were most frequently found in noncoding, intergenic regions of the rice genome. Microsatellites with poly(AT)n repeats represented the most abundant and polymorphic class of SSRs but were frequently associated with the Micropon family of miniature inverted-repeat transposable elements (MITEs) and were difficult to amplify. A set of 200 Class I SSR markers was developed and integrated into the existing microsatellite map of rice, providing immediate links between the genetic, physical, and sequence-based maps. This contribution brings the number of microsatellite markers that have been rigorously evaluated for amplification, map position, and allelic diversity in Oryza spp. to a total of 500.
Figures





Similar articles
-
Sequence variations of simple sequence repeats on chromosome-4 in two subspecies of the Asian cultivated rice.Theor Appl Genet. 2004 Feb;108(3):392-400. doi: 10.1007/s00122-003-1457-z. Epub 2003 Oct 17. Theor Appl Genet. 2004. PMID: 14564393
-
Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.).DNA Res. 2002 Dec 31;9(6):199-207. doi: 10.1093/dnares/9.6.199. DNA Res. 2002. PMID: 12597276
-
Nonrandom distribution and frequencies of genomic and EST-derived microsatellite markers in rice, wheat, and barley.BMC Genomics. 2005 Feb 18;6:23. doi: 10.1186/1471-2164-6-23. BMC Genomics. 2005. PMID: 15720707 Free PMC article.
-
Microsatellite marker development, mapping and applications in rice genetics and breeding.Plant Mol Biol. 1997 Sep;35(1-2):89-99. Plant Mol Biol. 1997. PMID: 9291963 Review.
-
Simple sequence repeat markers in genetic divergence and marker-assisted selection of rice cultivars: a review.Crit Rev Food Sci Nutr. 2015;55(1):41-9. doi: 10.1080/10408398.2011.646363. Crit Rev Food Sci Nutr. 2015. PMID: 24915404 Review.
Cited by
-
HighSSR: high-throughput SSR characterization and locus development from next-gen sequencing data.Bioinformatics. 2012 Nov 1;28(21):2797-803. doi: 10.1093/bioinformatics/bts524. Epub 2012 Sep 6. Bioinformatics. 2012. PMID: 22954626 Free PMC article.
-
Development of EST-SSR markers of Ipomoea nil.Breed Sci. 2012 Mar;62(1):99-104. doi: 10.1270/jsbbs.62.99. Epub 2012 Mar 20. Breed Sci. 2012. PMID: 23136520 Free PMC article.
-
Mining, characterization and validation of EST derived microsatellites from the transcriptome database of Allium sativum L.Bioinformation. 2015 Mar 31;11(3):145-50. doi: 10.6026/97320630011145. eCollection 2015. Bioinformation. 2015. PMID: 25987765 Free PMC article.
-
Patterned sequence in the transcriptome of vascular plants.BMC Genomics. 2007 Jun 15;8:173. doi: 10.1186/1471-2164-8-173. BMC Genomics. 2007. PMID: 17573970 Free PMC article.
-
Two complementary recessive genes in duplicated segments control etiolation in rice.Theor Appl Genet. 2011 Feb;122(2):373-83. doi: 10.1007/s00122-010-1453-z. Epub 2010 Sep 26. Theor Appl Genet. 2011. PMID: 20872210
References
-
- Akagi H, Yokozeki Y, Inagaki A, Fujimura T. Microsatellite DNA markers for rice chromosomes. Theor Appl Genet. 1996;94:61–67. - PubMed
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Aragon-Alcaide L, Miller T, Schwarzacher T, Reader S, Moore G. A cereal centromeric sequence. Chromosoma. 1996;105:261–268. - PubMed
-
- Arcot SS, Wan Z, Weber JL, Deininger PL, Batzer MA. Alu repeats: A source for the genesis of primate microsatellites. Genomics. 1995;29:136–144. - PubMed
-
- Bao JS, Zheng XW, Xia YW, He P, Shu QY, Lu X, Chen Y, Zhu LH. QTL mapping for the paste viscosity characteristics in rice (Oryza sativa L.) Theor Appl Genet. 2000;100:280–284.
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
