DNA looping in the RNA polymerase I enhancesome is the result of non-cooperative in-phase bending by two UBF molecules
- PMID: 11470882
- PMCID: PMC55825
- DOI: 10.1093/nar/29.15.3241
DNA looping in the RNA polymerase I enhancesome is the result of non-cooperative in-phase bending by two UBF molecules
Abstract
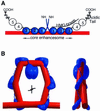
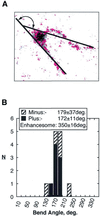
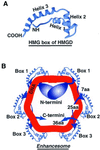
The so-called upstream binding factor (UBF) is required for the initial step in formation of an RNA polymerase I initiation complex. This function of UBF correlates with its ability to induce the ribosomal enhancesome, a structure which resembles in its mass and DNA content the nucleosome of chromatin. DNA looping in the enhancesome is probably the result of six in-phase bends induced by the HMG boxes of a UBF dimer. Here we show that insertion/deletion mutations in the basic peptide linker lying between the N-terminal dimerisation domain and the first HMG box of Xenopus UBF prevent the DNA looping characteristic of the enhancesome. Using these mutants we demonstrate that (i) the enhancesome structure does not depend on tethering of the entering and exiting DNA duplexes, (ii) UBF monomers induce hemi-enhancesomes, bending the DNA by 175 +/- 24 degrees and (iii) two hemi-enhancesomes are precisely phased by UBF dimerisation. We use this and previous data to refine the existing enhancesome model and show that HMG boxes 1 and 2 of UBF lie head-to-head along the DNA.
Figures


Similar articles
-
ERK modulates DNA bending and enhancesome structure by phosphorylating HMG1-boxes 1 and 2 of the RNA polymerase I transcription factor UBF.Biochemistry. 2006 Mar 21;45(11):3626-34. doi: 10.1021/bi051782h. Biochemistry. 2006. PMID: 16533045
-
The RNA polymerase I transcription factor UBF is a sequence-tolerant HMG-box protein that can recognize structured nucleic acids.Nucleic Acids Res. 1994 Jul 11;22(13):2651-7. doi: 10.1093/nar/22.13.2651. Nucleic Acids Res. 1994. PMID: 8041627 Free PMC article.
-
Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF.Science. 1994 May 20;264(5162):1134-7. doi: 10.1126/science.8178172. Science. 1994. PMID: 8178172
-
A role for upstream binding factor in organizing ribosomal gene chromatin.Biochem Soc Symp. 2006;(73):77-84. doi: 10.1042/bss0730077. Biochem Soc Symp. 2006. PMID: 16626289 Review.
-
Architectural transcription factors.Science. 1994 May 20;264(5162):1100-1. doi: 10.1126/science.8178167. Science. 1994. PMID: 8178167 Review. No abstract available.
Cited by
-
Involvement of a novel preimplantation-specific gene encoding the high mobility group box protein Hmgpi in early embryonic development.Hum Mol Genet. 2010 Feb 1;19(3):480-93. doi: 10.1093/hmg/ddp512. Epub 2009 Nov 14. Hum Mol Genet. 2010. PMID: 19915186 Free PMC article.
-
Regulation of Ribosome Biogenesis in Skeletal Muscle Hypertrophy.Physiology (Bethesda). 2019 Jan 1;34(1):30-42. doi: 10.1152/physiol.00034.2018. Physiology (Bethesda). 2019. PMID: 30540235 Free PMC article. Review.
-
A lattice model for transcription factor access to nucleosomal DNA.Biophys J. 2010 Oct 20;99(8):2597-607. doi: 10.1016/j.bpj.2010.08.019. Biophys J. 2010. PMID: 20959101 Free PMC article.
-
The DNA binding and bending activities of truncated tail-less HMGB1 protein are differentially affected by Lys-2 and Lys-81 residues and their acetylation.Int J Biol Sci. 2011;7(6):691-9. doi: 10.7150/ijbs.7.691. Epub 2011 Jun 1. Int J Biol Sci. 2011. PMID: 21647302 Free PMC article.
-
Perturbations at the ribosomal genes loci are at the centre of cellular dysfunction and human disease.Cell Biosci. 2014 Aug 19;4:43. doi: 10.1186/2045-3701-4-43. eCollection 2014. Cell Biosci. 2014. PMID: 25949792 Free PMC article. Review.
References
-
- Bazett-Jones D.P., Leblanc,B., Herfort,M. and Moss,T. (1994) Short-range DNA looping by the Xenopus HMG-box transcription factor, xUBF. Science, 264, 1134–1137. - PubMed
-
- Moss T. and Stefanovsky,V.Y. (1995) Promotion and regulation of ribosomal transcription in eukaryotes by RNA Polymerase I. In Cohn,W.E. and Moldave,K. (eds), Progress in Nucleic Acids and Molecular Biology. Academic Press, San Diego, CA, pp. 25–66. - PubMed
-
- Moss T., Stefanovsky,V.Y. and Pelletier,G. (1998) The structural and architectural role of Upstream Binding Factor, UBF. In Paule,M.R. (ed.), Transcription of Ribosomal Genes by Eukaryotic RNA Polymerase I. Landes Bioscience, Austin, TX, pp. 75–94.

