Complex formation by the human RAD51C and XRCC3 recombination repair proteins
- PMID: 11459987
- PMCID: PMC37455
- DOI: 10.1073/pnas.111005698
Complex formation by the human RAD51C and XRCC3 recombination repair proteins
Abstract
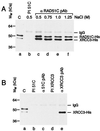
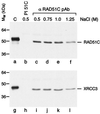
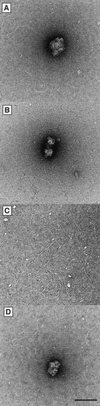
In vertebrates, the RAD51 protein is required for genetic recombination, DNA repair, and cellular proliferation. Five paralogs of RAD51, known as RAD51B, RAD51C, RAD51D, XRCC2, and XRCC3, have been identified and also shown to be required for recombination and genome stability. At the present time, however, very little is known about their biochemical properties or precise biological functions. As a first step toward understanding the roles of the RAD51 paralogs in recombination, the human RAD51C and XRCC3 proteins were overexpressed and purified from baculovirus-infected insect cells. The two proteins copurify as a complex, a property that reflects their endogenous association observed in HeLa cells. Purified RAD51C--XRCC3 complex binds single-stranded, but not duplex DNA, to form protein--DNA networks that have been visualized by electron microscopy.
Figures




Similar articles
-
Xrcc3 is recruited to DNA double strand breaks early and independent of Rad51.J Cell Biochem. 2004 Oct 15;93(3):429-36. doi: 10.1002/jcb.20232. J Cell Biochem. 2004. PMID: 15372620
-
Homologous-pairing activity of the human DNA-repair proteins Xrcc3.Rad51C.Proc Natl Acad Sci U S A. 2001 May 8;98(10):5538-43. doi: 10.1073/pnas.091603098. Epub 2001 May 1. Proc Natl Acad Sci U S A. 2001. PMID: 11331762 Free PMC article.
-
Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair.Plant J. 2005 Feb;41(4):533-45. doi: 10.1111/j.1365-313X.2004.02318.x. Plant J. 2005. PMID: 15686518
-
The RAD51 gene family, genetic instability and cancer.Cancer Lett. 2005 Mar 10;219(2):125-35. doi: 10.1016/j.canlet.2004.08.018. Cancer Lett. 2005. PMID: 15723711 Review.
-
RAD-ical New Insights into RAD51 Regulation.Genes (Basel). 2018 Dec 13;9(12):629. doi: 10.3390/genes9120629. Genes (Basel). 2018. PMID: 30551670 Free PMC article. Review.
Cited by
-
XRCC2 mutation causes meiotic arrest, azoospermia and infertility.J Med Genet. 2018 Sep;55(9):628-636. doi: 10.1136/jmedgenet-2017-105145. Epub 2018 Jul 24. J Med Genet. 2018. PMID: 30042186 Free PMC article.
-
The splicing-factor related protein SFPQ/PSF interacts with RAD51D and is necessary for homology-directed repair and sister chromatid cohesion.Nucleic Acids Res. 2011 Jan;39(1):132-45. doi: 10.1093/nar/gkq738. Epub 2010 Sep 2. Nucleic Acids Res. 2011. PMID: 20813759 Free PMC article.
-
Mechanistic Insights From Single-Molecule Studies of Repair of Double Strand Breaks.Front Cell Dev Biol. 2021 Nov 15;9:745311. doi: 10.3389/fcell.2021.745311. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34869333 Free PMC article. Review.
-
HELQ promotes RAD51 paralogue-dependent repair to avert germ cell loss and tumorigenesis.Nature. 2013 Oct 17;502(7471):381-4. doi: 10.1038/nature12565. Epub 2013 Sep 4. Nature. 2013. PMID: 24005329 Free PMC article.
-
A recombinase paralog from the hyperthermophilic crenarchaeon Sulfolobus solfataricus enhances SsoRadA ssDNA binding and strand displacement.Gene. 2013 Feb 15;515(1):128-39. doi: 10.1016/j.gene.2012.11.010. Epub 2012 Dec 6. Gene. 2013. PMID: 23220019 Free PMC article.
References
-
- McIlwraith M J, Van Dyck E, Masson J-Y, Stasiak A Z, Stasiak A, West S C. J Mol Biol. 2000;304:151–164. - PubMed
-
- Sung P. J Biol Chem. 1997;272:28194–28197. - PubMed
-
- Benson F E, Baumann P, West S C. Nature (London) 1998;391:401–404. - PubMed
-
- New J H, Sugiyama T, Zaitseva E, Kowalczykowski S C. Nature (London) 1998;391:407–410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

