Assembly of RecA-like recombinases: distinct roles for mediator proteins in mitosis and meiosis
- PMID: 11459983
- PMCID: PMC37451
- DOI: 10.1073/pnas.121046198
Assembly of RecA-like recombinases: distinct roles for mediator proteins in mitosis and meiosis
Abstract
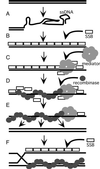
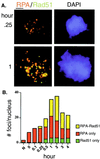
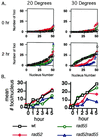
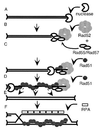
Members of the RecA family of recombinases from bacteriophage T4, Escherichia coli, yeast, and higher eukaryotes function in recombination as higher-order oligomers assembled on tracts of single-strand DNA (ssDNA). Biochemical studies have shown that assembly of recombinase involves accessory factors. These studies have identified a class of proteins, called recombination mediator proteins, that act by promoting assembly of recombinase on ssDNA tracts that are bound by ssDNA-binding protein (ssb). In the absence of mediators, ssb inhibits recombination reactions by competing with recombinase for DNA-binding sites. Here we briefly review mediated recombinase assembly and present results of new in vivo experiments. Immuno-double-staining experiments in Saccharomyces cerevisiae suggest that Rad51, the eukaryotic recombinase, can assemble at or near sites containing ssb (replication protein A, RPA) during the response to DNA damage, consistent with a need for mediator activity. Correspondingly, mediator gene mutants display defects in Rad51 assembly after DNA damage and during meiosis, although the requirements for assembly are distinct in the two cases. In meiosis, both Rad52 and Rad55/57 are required, whereas either Rad52 or Rad55/57 is sufficient to promote assembly of Rad51 in irradiated mitotic cells. Rad52 promotes normal amounts of Rad51 assembly in the absence of Rad55 at 30 degrees C but not 20 degrees C, accounting for the cold sensitivity of rad55 null mutants. Finally, we show that assembly of Rad51 is induced by radiation during S phase but not during G(1), consistent with the role of Rad51 in repairing the spontaneous damage that occurs during DNA replication.
Figures
Similar articles
-
Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase.Genes Dev. 1997 May 1;11(9):1111-21. doi: 10.1101/gad.11.9.1111. Genes Dev. 1997. PMID: 9159392
-
Rad52 associates with RPA and functions with rad55 and rad57 to assemble meiotic recombination complexes.Genes Dev. 1998 Jul 15;12(14):2208-21. doi: 10.1101/gad.12.14.2208. Genes Dev. 1998. PMID: 9679065 Free PMC article.
-
Stimulation by Rad52 of yeast Rad51-mediated recombination.Nature. 1998 Jan 22;391(6665):404-7. doi: 10.1038/34943. Nature. 1998. PMID: 9450759
-
Recombination proteins in yeast.Annu Rev Genet. 2004;38:233-71. doi: 10.1146/annurev.genet.38.072902.091500. Annu Rev Genet. 2004. PMID: 15568977 Review.
-
Role of the human RAD51 protein in homologous recombination and double-stranded-break repair.Trends Biochem Sci. 1998 Jul;23(7):247-51. doi: 10.1016/s0968-0004(98)01232-8. Trends Biochem Sci. 1998. PMID: 9697414 Review.
Cited by
-
In vivo assembly and disassembly of Rad51 and Rad52 complexes during double-strand break repair.EMBO J. 2004 Feb 25;23(4):939-49. doi: 10.1038/sj.emboj.7600091. Epub 2004 Feb 5. EMBO J. 2004. PMID: 14765116 Free PMC article.
-
Swi2/Snf2-related translocases prevent accumulation of toxic Rad51 complexes during mitotic growth.Mol Cell. 2010 Sep 24;39(6):862-72. doi: 10.1016/j.molcel.2010.08.028. Mol Cell. 2010. PMID: 20864034 Free PMC article.
-
Meiosis in budding yeast.Genetics. 2023 Oct 4;225(2):iyad125. doi: 10.1093/genetics/iyad125. Genetics. 2023. PMID: 37616582 Free PMC article.
-
RPA homologs and ssDNA processing during meiotic recombination.Chromosoma. 2016 Jun;125(2):265-76. doi: 10.1007/s00412-015-0552-7. Epub 2015 Oct 31. Chromosoma. 2016. PMID: 26520106 Free PMC article. Review.
-
DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination.Proc Natl Acad Sci U S A. 2001 Jul 17;98(15):8461-8. doi: 10.1073/pnas.151260698. Proc Natl Acad Sci U S A. 2001. PMID: 11459990 Free PMC article. Review.
References
-
- Howard F P, West S C, Stasiak A. Nature (London) 1984;309:215–219. - PubMed
-
- Stasiak A, Egelman E H. Experientia. 1994;50:192–203. - PubMed
-
- Griffith J, Formosa T. J Biol Chem. 1985;260:4484–4491. - PubMed
-
- Sung P, Robberson D L. Cell. 1995;82:453–461. - PubMed
-
- Ogawa T, Yu X, Shinohara A, Egelman E H. Science. 1993;259:1896–1899. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

