Communication between noncontacting macromolecules
- PMID: 11438725
- PMCID: PMC35404
- DOI: 10.1073/pnas.141221298
Communication between noncontacting macromolecules
Abstract
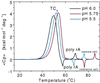
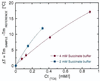
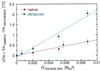
We present a quantitative experimental demonstration of solvent-mediated communication between noncontacting biopolymers. We show that changes in the activity of a solvent component brought about by a conformational change in one biopolymer can result in changes in the physical properties of a second noncontacting biopolymer present in solution. Specifically, we show that the release of protons on denaturation of a donor polymer (in this case, a four-stranded DNA tetraplex, iDNA) modulates the melting temperature of a noncontacting, acceptor polymer [in this case poly(A)]. In addition to such proton-mediated cross talk, we also demonstrate counterion-mediated cross talk between noncontacting biopolymers. Specifically, we show that counterion association/release on denaturation of native salmon sperm DNA (the donor polymer) can modulate the melting temperature of poly(dA) x poly(dT) (the acceptor polymer). Taken together, these two examples demonstrate how poly(A) and poly(dA) x poly(dT) can serve as molecular probes that report the pH and free salt concentrations in solution, respectively. Further, we demonstrate how such through-solvent dialogue between biopolymers that do not directly interact can be used to evaluate (in a model-free manner) association/dissociation reactions of solvent components (e.g., protons, sodium cations) with one of the two biopolymers. We propose that such through-solution dialogue is a general property of all biopolymers. As a result, such solvent-mediated cross talk should be considered when assessing reactions of multicomponent systems such as those that exist in essentially all biological processes.
Figures
Similar articles
-
Counterion association with native and denatured nucleic acids: an experimental approach.J Mol Biol. 2001 Jul 27;310(5):1011-25. doi: 10.1006/jmbi.2001.4841. J Mol Biol. 2001. PMID: 11501992
-
Communication between noncontacting macromolecules.Annu Rev Biophys Biomol Struct. 2005;34:21-42. doi: 10.1146/annurev.biophys.33.110502.133332. Annu Rev Biophys Biomol Struct. 2005. PMID: 15869382 Review.
-
Interaction of the alternating double stranded copolymer poly(dA-dT) x poly(dA-dT) with NiCl(2) and CdCl(2): solution behavior.J Biomol Struct Dyn. 2007 Aug;25(1):77-84. doi: 10.1080/07391102.2007.10507157. J Biomol Struct Dyn. 2007. PMID: 17676940
-
Binding of Escherichia coli primary replicative helicase DnaB protein to single-stranded DNA. Long-range allosteric conformational changes within the protein hexamer.Biochemistry. 1996 Feb 20;35(7):2129-45. doi: 10.1021/bi952345d. Biochemistry. 1996. PMID: 8652555
-
Conformational variability of poly(dA-dT).poly(dA-dT) and some other deoxyribonucleic acids includes a novel type of double helix.J Biomol Struct Dyn. 1985 Aug;3(1):67-83. doi: 10.1080/07391102.1985.10508399. J Biomol Struct Dyn. 1985. PMID: 3917211 Review.
Cited by
-
Aurones: small molecule visible range fluorescent probes suitable for biomacromolecules.J Fluoresc. 2011 Nov;21(6):2173-84. doi: 10.1007/s10895-011-0919-y. Epub 2011 Jul 12. J Fluoresc. 2011. PMID: 21748237 Free PMC article.
-
DNA repair and DNA triplet repeat expansion: the impact of abasic lesions on triplet repeat DNA energetics.J Am Chem Soc. 2009 Jul 8;131(26):9354-60. doi: 10.1021/ja902161e. J Am Chem Soc. 2009. PMID: 19566100 Free PMC article.
-
Energy landscapes of dynamic ensembles of rolling triplet repeat bulge loops: implications for DNA expansion associated with disease states.J Am Chem Soc. 2012 Apr 4;134(13):6033-44. doi: 10.1021/ja3010896. Epub 2012 Mar 23. J Am Chem Soc. 2012. PMID: 22397401 Free PMC article.
-
Energetic coupling between clustered lesions modulated by intervening triplet repeat bulge loops: allosteric implications for DNA repair and triplet repeat expansion.Biopolymers. 2010 Apr;93(4):355-69. doi: 10.1002/bip.21343. Biopolymers. 2010. PMID: 19890964 Free PMC article.
-
DNA meter: Energy tunable, quantitative hybridization assay.Biopolymers. 2013 Jun;99(6):408-17. doi: 10.1002/bip.22213. Biopolymers. 2013. PMID: 23529692 Free PMC article.
References
-
- Bloomfield V A, Crothers D M, Tinoco I., Jr . Nucleic Acids. Structures, Properties, and Functions. Mill Valley, CA: Univ. Sci. Books; 2000.
-
- Dobson C M, Sali A, Karplus M. Angew Chem Int Ed Engl. 1998;37:868–893. - PubMed
-
- Cantor C R, Schimmel P R. The Conformation of Biological Macromolecules. San Francisco: Freeman; 1980.
-
- Cantor C R, Schimmel P R. Techniques for the Study of Biological Structure and Function. San Francisco: Freeman; 1980.
-
- Cantor C R, Schimmel P R. The Behavior of Biological Macromolecules. San Francisco: Freeman; 1980.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

