Primer-dependent synthesis by poliovirus RNA-dependent RNA polymerase (3D(pol))
- PMID: 11433016
- PMCID: PMC55776
- DOI: 10.1093/nar/29.13.2715
Primer-dependent synthesis by poliovirus RNA-dependent RNA polymerase (3D(pol))
Abstract
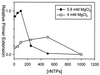
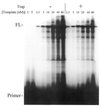
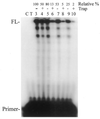
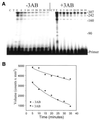
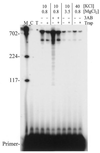
Properties of poliovirus RNA-dependent RNA polymerase (3D(pol)) including optimal conditions for primer extension, processivity and the rate of dissociation from primer-template (k(off)) were examined in the presence and absence of viral protein 3AB. Primer-dependent polymerization was examined on templates of 407 or 1499 nt primed such that fully extended products would be 296 or 1388 nt, respectively. Maximal primer extension was achieved with low rNTP concentrations (50-100 microM) using pH 7 and low (<1 mM) MgCl(2) and KCl (<20 mM) concentrations. However, high activity (about half maximal) was also observed with 500 microM rNTPs providing that higher MgCl(2) levels (3-5 mM) were used. The enhancement observed with the former conditions appeared to result from a large increase in the initial level or active enzyme that associated with the primer. 3AB increased the number of extended primers at all conditions with no apparent change in processivity. The k(off) values for the polymerase bound to primer-template were 0.011 +/- 0.005 and 0.037 +/- 0.006 min(-1) (average of four or more experiments +/- SD) in the presence or absence of 3AB, respectively. The decrease in the presence of 3AB suggested an enhancement of polymerase binding or stability. However, binding was tight even without 3AB, consistent with the highly processive (at least several hundred nucleotides) nature of 3D(pol). The results support a mechanism whereby 3AB enhances the ability of 3D(pol) to form a productive complex with the primer-template. Once formed, this complex is very stable resulting in highly processive synthesis.
Figures
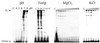
Similar articles
-
Characterization of protein-protein interactions critical for poliovirus replication: analysis of 3AB and VPg binding to the RNA-dependent RNA polymerase.J Virol. 2007 Jun;81(12):6369-78. doi: 10.1128/JVI.02252-06. Epub 2007 Apr 4. J Virol. 2007. PMID: 17409142 Free PMC article.
-
Poliovirus RNA-dependent RNA polymerase (3D(pol)). Divalent cation modulation of primer, template, and nucleotide selection.J Biol Chem. 1999 Dec 24;274(52):37060-9. doi: 10.1074/jbc.274.52.37060. J Biol Chem. 1999. PMID: 10601264
-
Intramolecular and intermolecular uridylylation by poliovirus RNA-dependent RNA polymerase.J Virol. 2006 Aug;80(15):7405-15. doi: 10.1128/JVI.02533-05. J Virol. 2006. PMID: 16840321 Free PMC article.
-
Picornavirus RNA polyadenylation by 3D(pol), the viral RNA-dependent RNA polymerase.Virus Res. 2015 Aug 3;206:3-11. doi: 10.1016/j.virusres.2014.12.030. Epub 2015 Jan 3. Virus Res. 2015. PMID: 25559071 Free PMC article. Review.
-
The Impact of Activating Agents on Non-Enzymatic Nucleic Acid Extension Reactions.Chembiochem. 2024 Apr 2;25(7):e202300859. doi: 10.1002/cbic.202300859. Epub 2024 Feb 12. Chembiochem. 2024. PMID: 38282207 Review.
Cited by
-
The twenty-nine amino acid C-terminal cytoplasmic domain of poliovirus 3AB is critical for nucleic acid chaperone activity.RNA Biol. 2010 Nov-Dec;7(6):820-9. doi: 10.4161/rna.7.6.13781. Epub 2010 Nov 1. RNA Biol. 2010. PMID: 21045553 Free PMC article.
-
Characterization of protein-protein interactions critical for poliovirus replication: analysis of 3AB and VPg binding to the RNA-dependent RNA polymerase.J Virol. 2007 Jun;81(12):6369-78. doi: 10.1128/JVI.02252-06. Epub 2007 Apr 4. J Virol. 2007. PMID: 17409142 Free PMC article.
-
Viral polymerases.Adv Exp Med Biol. 2012;726:267-304. doi: 10.1007/978-1-4614-0980-9_12. Adv Exp Med Biol. 2012. PMID: 22297518 Free PMC article. Review.
-
Purification and Biochemical Characterisation of Rabbit Calicivirus RNA-Dependent RNA Polymerases and Identification of Non-Nucleoside Inhibitors.Viruses. 2016 Apr 14;8(4):100. doi: 10.3390/v8040100. Viruses. 2016. PMID: 27089358 Free PMC article.
-
Poliovirus protein 3AB displays nucleic acid chaperone and helix-destabilizing activities.J Virol. 2006 Feb;80(4):1662-71. doi: 10.1128/JVI.80.4.1662-1671.2006. J Virol. 2006. PMID: 16439523 Free PMC article.
References
-
- Rueckert R.R. (1990) In Fields,B.N. and Knipe,D.M. (eds), Virology, 2nd Edn. Raven Press, New York, NY, Vol. 1, pp. 507–548.
-
- Houghton M. (1996) In Fields,B.N., Knipe,D.M. and Howley,P.M. (eds), Virology, 3rd Edn. Lippincott-Raven, Philidelphia, PA, Vol. 1, pp. 1035–1058.
-
- Neufeld K.L.R., Richards,O.C. and Ehrenfeld,E. (1991) Purification, characterization and comparison of poliovirus RNA polymerase from native and recombinant sources. J. Biol. Chem., 266, 24214–24219. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

