Repression of vaccinia virus Holliday junction resolvase inhibits processing of viral DNA into unit-length genomes
- PMID: 11413313
- PMCID: PMC114369
- DOI: 10.1128/JVI.75.14.6460-6471.2001
Repression of vaccinia virus Holliday junction resolvase inhibits processing of viral DNA into unit-length genomes
Abstract
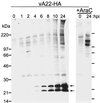
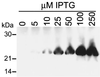
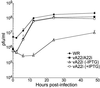
The vaccinia virus A22R gene encodes a protein that is homologous to the bacterial enzyme RuvC and specifically cleaves and resolves four-way DNA Holliday junctions into linear duplex products. To investigate the role of the vaccinia virus Holliday junction resolvase during an infection, we constructed two recombinant viruses: vA22-HA, which has a short C-terminal epitope tag appended to the A22R open reading frame, and vA22i, in which the original A22R gene is deleted and replaced by an inducible copy. Polyacrylamide gel electrophoresis and Western blot analysis of extracts and purified virions from cells infected with vA22-HA revealed that the resolvase was expressed after the onset of DNA replication and incorporated into virion cores. vA22i exhibited a conditional replication defect. In the absence of an inducer, (i) viral protein synthesis was unaffected, (ii) late-stage viral DNA replication was reduced, (iii) most of the newly synthesized viral DNA remained in a branched or concatemeric form that caused it to be trapped at the application site during pulsed-field gel electrophoresis, (iv) cleavage of concatemer junctions was inhibited, and (v) virion morphogenesis was arrested at an immature stage. These data indicated multiple roles for the vaccinia virus Holliday junction resolvase in the replication and processing of viral DNA into unit-length genomes.
Figures






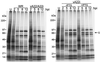

Similar articles
-
DNA binding and cleavage by the fowlpox virus resolvase.J Biol Chem. 2009 Jan 9;284(2):1190-201. doi: 10.1074/jbc.M807864200. Epub 2008 Nov 12. J Biol Chem. 2009. PMID: 19004818 Free PMC article.
-
DNA cleavage by the A22R resolvase of vaccinia virus.Virology. 2006 Sep 1;352(2):466-76. doi: 10.1016/j.virol.2006.05.007. Epub 2006 Jun 15. Virology. 2006. PMID: 16781759
-
Vaccinia virus mutants with alanine substitutions in the conserved G5R gene fail to initiate morphogenesis at the nonpermissive temperature.J Virol. 2004 Oct;78(19):10238-48. doi: 10.1128/JVI.78.19.10238-10248.2004. J Virol. 2004. PMID: 15367589 Free PMC article.
-
Vaccinia virus WR gene A5L is required for morphogenesis of mature virions.J Virol. 1999 Jun;73(6):4590-9. doi: 10.1128/JVI.73.6.4590-4599.1999. J Virol. 1999. PMID: 10233918 Free PMC article.
-
Vaccinia virus G1 protein, a predicted metalloprotease, is essential for morphogenesis of infectious virions but not for cleavage of major core proteins.J Virol. 2004 Jul;78(13):6855-63. doi: 10.1128/JVI.78.13.6855-6863.2004. J Virol. 2004. PMID: 15194761 Free PMC article.
Cited by
-
Generation of a complete single-gene knockout bacterial artificial chromosome library of cowpox virus and identification of its essential genes.J Virol. 2014 Jan;88(1):490-502. doi: 10.1128/JVI.02385-13. Epub 2013 Oct 23. J Virol. 2014. PMID: 24155400 Free PMC article.
-
Inhibition of the ubiquitin-proteasome system prevents vaccinia virus DNA replication and expression of intermediate and late genes.J Virol. 2009 Mar;83(6):2469-79. doi: 10.1128/JVI.01986-08. Epub 2009 Jan 7. J Virol. 2009. PMID: 19129442 Free PMC article.
-
DNA binding and cleavage by the fowlpox virus resolvase.J Biol Chem. 2009 Jan 9;284(2):1190-201. doi: 10.1074/jbc.M807864200. Epub 2008 Nov 12. J Biol Chem. 2009. PMID: 19004818 Free PMC article.
-
Products and substrate/template usage of vaccinia virus DNA primase.Virology. 2009 Jan 5;383(1):136-41. doi: 10.1016/j.virol.2008.10.008. Epub 2008 Nov 12. Virology. 2009. PMID: 19007959 Free PMC article.
-
Low-resolution structure of vaccinia virus DNA replication machinery.J Virol. 2013 Feb;87(3):1679-89. doi: 10.1128/JVI.01533-12. Epub 2012 Nov 21. J Virol. 2013. PMID: 23175373 Free PMC article.
References
-
- Baroudy B M, Venkatesan S, Moss B. Incompletely base-paired flip-flop terminal loops link the two DNA strands of the vaccinia virus genome into one uninterrupted polynucleotide chain. Cell. 1982;28:315–324. - PubMed
-
- Baroudy B M, Venkatesan S, Moss B. Structure and replication of vaccinia virus telomeres. Cold Spring Harbor Symp Quant Biol. 1982;47:723–729. - PubMed
-
- Black L W. DNA packaging and cutting by phage terminases: control in phage T4 by a synaptic mechanism. Bioessays. 1995;17:1025–1030. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

