Issues in cDNA microarray analysis: quality filtering, channel normalization, models of variations and assessment of gene effects
- PMID: 11410663
- PMCID: PMC55725
- DOI: 10.1093/nar/29.12.2549
Issues in cDNA microarray analysis: quality filtering, channel normalization, models of variations and assessment of gene effects
Abstract
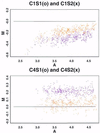
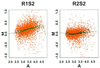
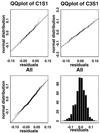
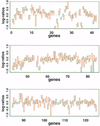
We consider the problem of comparing the gene expression levels of cells grown under two different conditions using cDNA microarray data. We use a quality index, computed from duplicate spots on the same slide, to filter out outlying spots, poor quality genes and problematical slides. We also perform calibration experiments to show that normalization between fluorescent labels is needed and that the normalization is slide dependent and non-linear. A rank invariant method is suggested to select non-differentially expressed genes and to construct normalization curves in comparative experiments. After normalization the residuals from the calibration data are used to provide prior information on variance components in the analysis of comparative experiments. Based on a hierarchical model that incorporates several levels of variations, a method for assessing the significance of gene effects in comparative experiments is presented. The analysis is demonstrated via two groups of experiments with 125 and 4129 genes, respectively, in Escherichia coli grown in glucose and acetate.
Figures




Similar articles
-
Model selection and efficiency testing for normalization of cDNA microarray data.Genome Biol. 2004;5(8):R60. doi: 10.1186/gb-2004-5-8-r60. Epub 2004 Jul 30. Genome Biol. 2004. PMID: 15287982 Free PMC article.
-
Effect of local background intensities in the normalization of cDNA microarray data with a skewed expression profiles.Exp Mol Med. 2002 Jul 31;34(3):224-32. doi: 10.1038/emm.2002.31. Exp Mol Med. 2002. PMID: 12216114
-
Assessment of 35mer amino-modified oligonucleotide based microarray with bacterial samples.J Microbiol Methods. 2004 May;57(2):207-18. doi: 10.1016/j.mimet.2004.01.009. J Microbiol Methods. 2004. PMID: 15063061
-
Normalization and quantification of differential expression in gene expression microarrays.Brief Bioinform. 2006 Jun;7(2):166-77. doi: 10.1093/bib/bbl002. Epub 2006 Mar 7. Brief Bioinform. 2006. PMID: 16772260 Review.
-
Standards in gene expression microarray experiments.Methods Enzymol. 2006;411:63-78. doi: 10.1016/S0076-6879(06)11005-8. Methods Enzymol. 2006. PMID: 16939786 Review.
Cited by
-
Normalization and analysis of cDNA microarrays using within-array replications applied to neuroblastoma cell response to a cytokine.Proc Natl Acad Sci U S A. 2004 Feb 3;101(5):1135-40. doi: 10.1073/pnas.0307557100. Epub 2004 Jan 22. Proc Natl Acad Sci U S A. 2004. PMID: 14739336 Free PMC article.
-
A novel normalization method for effective removal of systematic variation in microarray data.Nucleic Acids Res. 2006 Mar 9;34(5):e38. doi: 10.1093/nar/gkl024. Print 2006. Nucleic Acids Res. 2006. PMID: 16528099 Free PMC article.
-
Conserved and variable functions of the sigmaE stress response in related genomes.PLoS Biol. 2006 Jan;4(1):e2. doi: 10.1371/journal.pbio.0040002. PLoS Biol. 2006. PMID: 16336047 Free PMC article.
-
Subset quantile normalization using negative control features.J Comput Biol. 2010 Oct;17(10):1385-95. doi: 10.1089/cmb.2010.0049. J Comput Biol. 2010. PMID: 20976876 Free PMC article.
-
BeadDataPackR: A Tool to Facilitate the Sharing of Raw Data from Illumina BeadArray Studies.Cancer Inform. 2010 Sep 29;9:217-27. Cancer Inform. 2010. PMID: 20981138 Free PMC article.
References
-
- Brown P.O. and Botstein,D. (1999) Exploring the new world of the genome with DNA microarrays. Nat. Genet., 21 (suppl. 1), 33–37. - PubMed
-
- Chen Y., Dougherty,E.R. and Bittner,M.L. (1997) Ratio-based decisions and the quantitative analysis of cDNA microarray images. J. Biomed. Optics, 2, 364–374. - PubMed
-
- Dudoit Y., Yang,Y.H., Callow,M.J. and Speed,T.P. (2000) Statistical methods for identifying differentially expressed genes in replicated cDNA microarray experiments. Technical Report 578, Department of Statistics, UC Berkeley, CA.
-
- Kerr M.K. and Churchill,G.A. (2000) Experimental design for gene expression microarrays. Biostatistics, in press. - PubMed
-
- Kerr M.K., Martin,M. and Churchill,G.A. (2000) Analysis of variance for gene expression microarray. J. Comput. Biol., 7, 819–837. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

