Pneumococcal virulence factors: structure and function
- PMID: 11381099
- PMCID: PMC99024
- DOI: 10.1128/MMBR.65.2.187-207.2001
Pneumococcal virulence factors: structure and function
Abstract
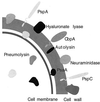
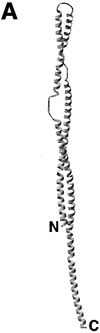
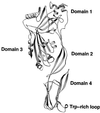
The overall goal for this review is to summarize the current body of knowledge about the structure and function of major known antigens of Streptococcus pneumoniae, a major gram-positive bacterial pathogen of humans. This information is then related to the role of these proteins in pneumococcal pathogenesis and in the development of new vaccines and/or other antimicrobial agents. S. pneumoniae is the most common cause of fatal community-acquired pneumonia in the elderly and is also one of the most common causes of middle ear infections and meningitis in children. The present vaccine for the pneumococcus consists of a mixture of 23 different capsular polysaccharides. While this vaccine is very effective in young adults, who are normally at low risk of serious disease, it is only about 60% effective in the elderly. In children younger than 2 years the vaccine is ineffective and is not recommended due to the inability of this age group to mount an antibody response to the pneumococcal polysaccharides. Antimicrobial drugs such as penicillin have diminished the risk from pneumococcal disease. Several pneumococcal proteins including pneumococcal surface proteins A and C, hyaluronate lyase, pneumolysin, autolysin, pneumococcal surface antigen A, choline binding protein A, and two neuraminidase enzymes are being investigated as potential vaccine or drug targets. Essentially all of these antigens have been or are being investigated on a structural level in addition to being characterized biochemically. Recently, three-dimensional structures for hyaluronate lyase and pneumococcal surface antigen A became available from X-ray crystallography determinations. Also, modeling studies based on biophysical measurements provided more information about the structures of pneumolysin and pneumococcal surface protein A. Structural and biochemical studies of these pneumococcal virulence factors have facilitated the development of novel antibiotics or protein antigen-based vaccines as an alternative to polysaccharide-based vaccines for the treatment of pneumococcal disease.
Figures






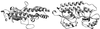

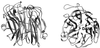
Similar articles
-
[Proteins of Streptococcus pneumoniae: perspectives for development of pneumococcal vaccine].Zh Mikrobiol Epidemiol Immunobiol. 2010 Nov-Dec;(6):98-104. Zh Mikrobiol Epidemiol Immunobiol. 2010. PMID: 21384595 Review. Russian.
-
Molecular analysis of the pathogenicity of Streptococcus pneumoniae: the role of pneumococcal proteins.Annu Rev Microbiol. 1993;47:89-115. doi: 10.1146/annurev.mi.47.100193.000513. Annu Rev Microbiol. 1993. PMID: 7903033 Review.
-
Molecular and cellular biology of pneumococcal infection.Curr Opin Microbiol. 1999 Feb;2(1):35-9. doi: 10.1016/s1369-5274(99)80006-x. Curr Opin Microbiol. 1999. PMID: 10047549 Review.
-
Pneumolysin as a vaccine and drug target in the prevention and treatment of invasive pneumococcal disease.Arch Immunol Ther Exp (Warsz). 2005 May-Jun;53(3):189-98. Arch Immunol Ther Exp (Warsz). 2005. PMID: 15995579 Review.
-
The role of pneumolysin in the pathogenesis of Streptococcus pneumoniae infection.Curr Opin Infect Dis. 2002 Jun;15(3):235-9. doi: 10.1097/00001432-200206000-00004. Curr Opin Infect Dis. 2002. PMID: 12015456 Review.
Cited by
-
CrfP, a fratricide protein, contributes to natural transformation in Streptococcus suis.Vet Res. 2021 Mar 24;52(1):50. doi: 10.1186/s13567-021-00917-x. Vet Res. 2021. PMID: 33762005 Free PMC article.
-
Streptococcal phosphotransferase system imports unsaturated hyaluronan disaccharide derived from host extracellular matrices.PLoS One. 2019 Nov 7;14(11):e0224753. doi: 10.1371/journal.pone.0224753. eCollection 2019. PLoS One. 2019. PMID: 31697725 Free PMC article.
-
SecA2-dependent secretion of autolytic enzymes promotes Listeria monocytogenes pathogenesis.Proc Natl Acad Sci U S A. 2003 Oct 14;100(21):12432-7. doi: 10.1073/pnas.2133653100. Epub 2003 Oct 3. Proc Natl Acad Sci U S A. 2003. PMID: 14527997 Free PMC article.
-
Purification and preliminary crystallization of alanine racemase from Streptococcus pneumoniae.BMC Microbiol. 2007 May 17;7:40. doi: 10.1186/1471-2180-7-40. BMC Microbiol. 2007. PMID: 17509154 Free PMC article.
-
Immunization with native or recombinant Streptococcus pneumoniae neuraminidase affords protection in the chinchilla otitis media model.Infect Immun. 2004 Jul;72(7):4309-13. doi: 10.1128/IAI.72.7.4309-4313.2004. Infect Immun. 2004. PMID: 15213181 Free PMC article.
References
-
- Afessa B, Greaves W L, Frederick W R. Pneumococcal bacteremia in adults: a 14-year experience in an inner-city university hospital. Clin Infect Dis. 1995;21:345–351. - PubMed
-
- Arnold K E, Leggiadro R J, Breiman R F, et al. Risk factors for carriage of drug-resistant Streptococcus pneumoniae among children in Memphis, Tennessee. J Pediatr. 1996;128:757–764. - PubMed
-
- Aruffo A, Stamenkovic I, Melnick M, Underhill C B, Seed B. CD44 is the principal cell surface receptor for hyaluronan. Cell. 1990;61:1303–1313. - PubMed
-
- Austrian R. Some observations on the pneumococcus and on the current status of pneumococcal disease and its prevention. In: Quie P G, Kass E H, editors. The pneumococcus and the pneumococcal vaccine. Chicago, Ill: University of Chicago Press; 1982. pp. 191–207.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

