Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription
- PMID: 11350943
- PMCID: PMC125250
- DOI: 10.1093/emboj/20.10.2536
Dnmt3a binds deacetylases and is recruited by a sequence-specific repressor to silence transcription
Abstract
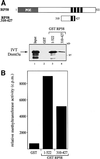
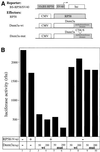
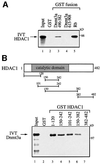
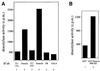
The Dnmt3a DNA methyltransferase is essential for mammalian development and is responsible for the generation of genomic methylation patterns, which lead to transcriptional silencing. Here, we show that Dnmt3a associates with RP58, a DNA-binding transcriptional repressor protein found at transcriptionally silent heterochromatin. Dnmt3a acts as a co-repressor for RP58 in a manner that does not require its de novo methyltransferase activity. Like other characterized co-repressors, Dnmt3a associates with the histone deacetylase HDAC1 using its ATRX-homology domain. This domain of Dnmt3a represents an independent transcriptional repressor domain whose silencing functions require HDAC activity. These results identify Dnmt3a as a co-repressor protein carrying deacetylase activity and show that Dnmt3a can be targeted to specific regulatory foci via its association with DNA-binding transcription factors.
Figures




Comment in
-
A catalytic dependent role for DNMT3B in tumor suppression.EBioMedicine. 2021 Mar;65:103237. doi: 10.1016/j.ebiom.2021.103237. Epub 2021 Mar 11. EBioMedicine. 2021. PMID: 33714886 Free PMC article. No abstract available.
Similar articles
-
Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin.J Biol Chem. 2001 Aug 24;276(34):32282-7. doi: 10.1074/jbc.M104661200. Epub 2001 Jun 26. J Biol Chem. 2001. PMID: 11427539
-
Imprinting regulator DNMT3L is a transcriptional repressor associated with histone deacetylase activity.Nucleic Acids Res. 2002 Aug 15;30(16):3602-8. doi: 10.1093/nar/gkf474. Nucleic Acids Res. 2002. PMID: 12177302 Free PMC article.
-
Physical and functional interaction of DNA methyltransferase 3A with Mbd3 and Brg1 in mouse lymphosarcoma cells.Cancer Res. 2005 Dec 1;65(23):10891-900. doi: 10.1158/0008-5472.CAN-05-1455. Cancer Res. 2005. Retraction in: Cancer Res. 2023 Dec 15;83(24):4182. doi: 10.1158/0008-5472.CAN-23-2344 PMID: 16322236 Free PMC article. Retracted.
-
Histone deacetylase directs the dominant silencing of transcription in chromatin: association with MeCP2 and the Mi-2 chromodomain SWI/SNF ATPase.Cold Spring Harb Symp Quant Biol. 1998;63:435-45. doi: 10.1101/sqb.1998.63.435. Cold Spring Harb Symp Quant Biol. 1998. PMID: 10384308 Review. No abstract available.
-
Modulation of thyroid hormone receptor silencing function by co-repressors and a synergizing transcription factor.Biochem Soc Trans. 2000;28(4):386-9. Biochem Soc Trans. 2000. PMID: 10961925 Review.
Cited by
-
DNA methyltransferases in hematological malignancies.J Genet Genomics. 2020 Jul 20;47(7):361-372. doi: 10.1016/j.jgg.2020.04.006. Epub 2020 Jul 24. J Genet Genomics. 2020. PMID: 32994141 Free PMC article. Review.
-
Enzymology of Mammalian DNA Methyltransferases.Adv Exp Med Biol. 2022;1389:69-110. doi: 10.1007/978-3-031-11454-0_4. Adv Exp Med Biol. 2022. PMID: 36350507
-
Allosteric control of mammalian DNA methyltransferases - a new regulatory paradigm.Nucleic Acids Res. 2016 Oct 14;44(18):8556-8575. doi: 10.1093/nar/gkw723. Epub 2016 Aug 12. Nucleic Acids Res. 2016. PMID: 27521372 Free PMC article. Review.
-
Molecular and enzymatic profiles of mammalian DNA methyltransferases: structures and targets for drugs.Curr Med Chem. 2010;17(33):4052-71. doi: 10.2174/092986710793205372. Curr Med Chem. 2010. PMID: 20939822 Free PMC article. Review.
-
Epigenetic control of nuclear architecture.Cell Mol Life Sci. 2007 Feb;64(4):449-57. doi: 10.1007/s00018-007-6358-x. Cell Mol Life Sci. 2007. PMID: 17221334 Free PMC article. Review.
References
-
- Aapola U. et al. (2000) Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics, 65, 293–298. - PubMed
-
- Aasland R., Gibson,T.J. and Stewart,A.F. (1995) The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci., 20, 56–59. - PubMed
-
- Aoki K., Meng,G., Suzuki,K., Takashi,T., Kameoka,Y., Nakahara,K., Ishida,R. and Kasai,M. (1998) RP58 associates with condensed chromatin and mediates a sequence- specific transcriptional repression. J. Biol. Chem., 273, 26698–26704. - PubMed
-
- Bannister A.J. and Kouzarides,T. (1996) The CBP co-activator is a histone acetyltransferase. Nature, 384, 641–643. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

