Identification and characterization of a second superoxide dismutase gene (sodM) from Staphylococcus aureus
- PMID: 11344148
- PMCID: PMC99638
- DOI: 10.1128/JB.183.11.3399-3407.2001
Identification and characterization of a second superoxide dismutase gene (sodM) from Staphylococcus aureus
Abstract
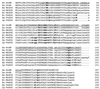
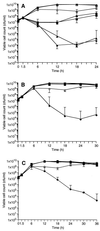
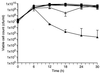
A gene encoding superoxide dismutase (SOD), sodM, from S. aureus was cloned and characterized. The deduced amino acid sequence specifies a 187-amino-acid protein with 75% identity to the S. aureus SodA protein. Amino acid sequence comparisons with known SODs and relative insensitivity to hydrogen peroxide and potassium cyanide indicate that SodM most likely uses manganese (Mn) as a cofactor. The sodM gene expressed from a plasmid rescued an Escherichia coli double mutant (sodA sodB) under conditions that are otherwise lethal. SOD activity gels of S. aureus RN6390 whole-cell lysates revealed three closely migrating bands of activity. The two upper bands were absent in a sodM mutant, while the two lower bands were absent in a sodA mutant. Thus, the middle band of activity most likely represents a SodM-SodA hybrid protein. All three bands of activity increased as highly aerated cultures entered the late exponential phase of growth, SodM more so than SodA. Viability of the sodA and sodM sodA mutants but not the sodM mutant was drastically reduced under oxidative stress conditions generated by methyl viologen (MV) added during the early exponential phase of growth. However, only the viability of the sodM sodA mutant was reduced when MV was added during the late exponential and stationary phases of growth. These data indicate that while SodA may be the major SOD activity in S. aureus throughout all stages of growth, SodM, under oxidative stress, becomes a major source of activity during the late exponential and stationary phases of growth such that viability and growth of an S. aureus sodA mutant are maintained.
Figures

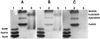
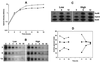
Similar articles
-
Regulation of superoxide dismutase (sod) genes by SarA in Staphylococcus aureus.J Bacteriol. 2009 May;191(10):3301-10. doi: 10.1128/JB.01496-08. Epub 2009 Mar 13. J Bacteriol. 2009. PMID: 19286803 Free PMC article.
-
The superoxide dismutase gene sodM is unique to Staphylococcus aureus: absence of sodM in coagulase-negative staphylococci.J Bacteriol. 2002 May;184(9):2465-72. doi: 10.1128/JB.184.9.2465-2472.2002. J Bacteriol. 2002. PMID: 11948161 Free PMC article.
-
Role and regulation of the superoxide dismutases of Staphylococcus aureus.Microbiology (Reading). 2003 Oct;149(Pt 10):2749-2758. doi: 10.1099/mic.0.26353-0. Microbiology (Reading). 2003. PMID: 14523108
-
Pseudomonas aeruginosa sodA and sodB mutants defective in manganese- and iron-cofactored superoxide dismutase activity demonstrate the importance of the iron-cofactored form in aerobic metabolism.J Bacteriol. 1995 Nov;177(22):6330-7. doi: 10.1128/jb.177.22.6330-6337.1995. J Bacteriol. 1995. PMID: 7592406 Free PMC article.
-
The molecular genetics of superoxide dismutase in E. coli. An approach to understanding the biological role and regulation of SODS in relation to other elements of the defence system against oxygen toxicity.Free Radic Res Commun. 1989;8(1):1-9. doi: 10.3109/10715768909087967. Free Radic Res Commun. 1989. PMID: 2684801 Review.
Cited by
-
Regulation of superoxide dismutase (sod) genes by SarA in Staphylococcus aureus.J Bacteriol. 2009 May;191(10):3301-10. doi: 10.1128/JB.01496-08. Epub 2009 Mar 13. J Bacteriol. 2009. PMID: 19286803 Free PMC article.
-
Superoxide dismutase is upregulated in Staphylococcus aureus following protoporphyrin-mediated photodynamic inactivation and does not directly influence the response to photodynamic treatment.BMC Microbiol. 2010 Dec 17;10:323. doi: 10.1186/1471-2180-10-323. BMC Microbiol. 2010. PMID: 21167031 Free PMC article.
-
Comparative Proteomic and Morphological Change Analyses of Staphylococcus aureus During Resuscitation From Prolonged Freezing.Front Microbiol. 2018 May 3;9:866. doi: 10.3389/fmicb.2018.00866. eCollection 2018. Front Microbiol. 2018. PMID: 29774015 Free PMC article.
-
Exploring the targetome of IsrR, an iron-regulated sRNA controlling the synthesis of iron-containing proteins in Staphylococcus aureus.Front Microbiol. 2024 Jul 5;15:1439352. doi: 10.3389/fmicb.2024.1439352. eCollection 2024. Front Microbiol. 2024. PMID: 39035440 Free PMC article.
-
Regulation of Staphylococcus aureus MntC expression and its role in response to oxidative stress.PLoS One. 2013 Oct 28;8(10):e77874. doi: 10.1371/journal.pone.0077874. eCollection 2013. PLoS One. 2013. PMID: 24205007 Free PMC article.
References
-
- Asada K, Yoshikawa K, Takahashi M-A, Maeda Y, Enmanji K. Superoxide dismutases from a blue-green alga, Plectonema boryanum. J Biol Chem. 1975;250:2801–2807. - PubMed
-
- Beauchamp C, Fridovich I. Superoxide dismutase: improved assays and an assay applicable to acrylamide gels. Anal Biochem. 1971;44:276–287. - PubMed
-
- Blevins J S, Gillaspy A F, Rechtin T M, Hurlburt B K, Smeltzer M S. The staphylococcal accessory regulator (sar) represses transcription of the Staphylococcus aureus collagen adhesin gene (cna) in an agr-independent manner. Mol Microbiol. 1999;33:317–326. - PubMed
-
- Centers for Disease Control and Prevention. National nosocomial infection surveillance system report: data summary from October 1986–April 1996. U.S. Atlanta, Ga: Department of Health and Human Services; 1996.
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

