Mutations in the novel membrane protein spinster interfere with programmed cell death and cause neural degeneration in Drosophila melanogaster
- PMID: 11340170
- PMCID: PMC87027
- DOI: 10.1128/MCB.21.11.3775-3788.2001
Mutations in the novel membrane protein spinster interfere with programmed cell death and cause neural degeneration in Drosophila melanogaster
Abstract
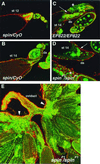
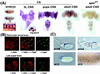
Mutations in the spin gene are characterized by an extraordinarily strong rejection behavior of female flies in response to male courtship. They are also accompanied by decreases in the viability, adult life span, and oviposition rate of the flies. In spin mutants, some oocytes and adult neural cells undergo degeneration, which is preceded by reductions in programmed cell death of nurse cells in ovaries and of neurons in the pupal nervous system, respectively. The central nervous system (CNS) of spin mutant flies accumulates autofluorescent lipopigments with characteristics similar to those of lipofuscin. The spin locus generates at least five different transcripts, with only two of these being able to rescue the spin behavioral phenotype; each encodes a protein with multiple membrane-spanning domains that are expressed in both the surface glial cells in the CNS and the follicle cells in the ovaries. Orthologs of the spin gene have also been identified in a number of species from nematodes to humans. Analysis of the spin mutant will give us new insights into neurodegenerative diseases and aging.
Figures


Similar articles
-
Pathology of the adult central nervous system induced by genetic inhibition of programmed cell death in Drosophila pupae.Arch Insect Biochem Physiol. 2002 Feb;49(2):94-101. doi: 10.1002/arch.10011. Arch Insect Biochem Physiol. 2002. PMID: 11816024
-
Zebrafish yolk-specific not really started (nrs) gene is a vertebrate homolog of the Drosophila spinster gene and is essential for embryogenesis.Dev Dyn. 2002 Mar;223(2):298-305. doi: 10.1002/dvdy.10060. Dev Dyn. 2002. PMID: 11836794
-
Phenotypic interactions of spinster with the genes encoding proteins for cell death control in Drosophila melanogaster.Arch Insect Biochem Physiol. 2010 Mar;73(3):119-27. doi: 10.1002/arch.20345. Arch Insect Biochem Physiol. 2010. PMID: 20091795
-
Phagocyte Responses to Cell Death in Flies.Cold Spring Harb Perspect Biol. 2020 Apr 1;12(4):a036350. doi: 10.1101/cshperspect.a036350. Cold Spring Harb Perspect Biol. 2020. PMID: 31501193 Free PMC article. Review.
-
Tumor suppressor genes and signal transduction in Drosophila.Princess Takamatsu Symp. 1994;24:1-13. Princess Takamatsu Symp. 1994. PMID: 8983059 Review.
Cited by
-
Neurodegenerative mutants in Drosophila: a means to identify genes and mechanisms involved in human diseases?Invert Neurosci. 2005 Nov;5(3-4):97-109. doi: 10.1007/s10158-005-0005-8. Epub 2005 Oct 24. Invert Neurosci. 2005. PMID: 16187075 Review.
-
Genetic modifiers of the Drosophila blue cheese gene link defects in lysosomal transport with decreased life span and altered ubiquitinated-protein profiles.Genetics. 2007 Jun;176(2):1283-97. doi: 10.1534/genetics.106.065011. Epub 2007 Apr 15. Genetics. 2007. PMID: 17435236 Free PMC article.
-
blue cheese mutations define a novel, conserved gene involved in progressive neural degeneration.J Neurosci. 2003 Feb 15;23(4):1254-64. doi: 10.1523/JNEUROSCI.23-04-01254.2003. J Neurosci. 2003. PMID: 12598614 Free PMC article.
-
A neural circuit encoding mating states tunes defensive behavior in Drosophila.Nat Commun. 2020 Aug 7;11(1):3962. doi: 10.1038/s41467-020-17771-8. Nat Commun. 2020. PMID: 32770059 Free PMC article.
-
The nuclear protein Waharan is required for endosomal-lysosomal trafficking in Drosophila.J Cell Sci. 2010 Jul 15;123(Pt 14):2369-74. doi: 10.1242/jcs.060582. Epub 2010 Jun 15. J Cell Sci. 2010. PMID: 20551180 Free PMC article.
References
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Becker L E, Prior T W, Yates A J. Metabolic disease. In: Davis R L, Robertson D M, editors. Textbook of neuropathology. Baltimore, Md: The Williams & Willkins Co.; 1997. pp. 407–509.
-
- Brand A H, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. - PubMed
-
- Buchanan R L, Benzer S. Defective glia in the Drosophila brain degeneration mutant drop-dead. Neuron. 1993;10:839–850. - PubMed
-
- Buege J A, Aust S D. Microsomal lipid peroxidation. Methods Enzymol. 1978;52:302–310. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
