Virological properties and nucleotide sequences of Cas-E-type endogenous ecotropic murine leukemia viruses in South Asian wild mice, Mus musculus castaneus
- PMID: 11333885
- PMCID: PMC114909
- DOI: 10.1128/JVI.75.11.5049-5058.2001
Virological properties and nucleotide sequences of Cas-E-type endogenous ecotropic murine leukemia viruses in South Asian wild mice, Mus musculus castaneus
Abstract
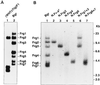
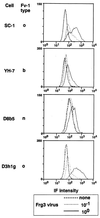
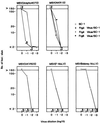
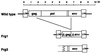
Two types of endogenous ecotropic murine leukemia viruses (MuLVs), termed AKV- and Cas-E-type MuLVs, differ in nucleotide sequence and distribution in wild mouse subspecies. In contrast to AKV-type MuLV, Cas-E-type MuLV is not carried by common laboratory mice. Wild mice of Mus musculus (M. m.) castaneus carry multiple copies of Cas-E-type endogenous MuLV, including the Fv-4(r) gene that is a truncated form of integrated MuLV and functions as a host's resistance gene against ecotropic MuLV infection. Our genetic cross experiments showed that only the Fv-4(r) gene was associated with resistance to ecotropic F-MuLV infection. Because the spontaneous expression of infectious virus was not detected in M. m. castaneus, we generated mice that did not carry the Fv-4(r) gene but did carry a single or a few endogenous MuLV loci. In mice not carrying the Fv-4(r) gene, infectious MuLVs were isolated in association with three of six Cas-E-type endogenous MuLV loci. The isolated viruses showed a weak syncytium-forming activity for XC cells, an interfering property of ecotropic MuLV, and a slight antigenic variation. Two genomic DNAs containing endogenous Cas-E-type MuLV were cloned and partially sequenced. All of the Cas-E-type endogenous MuLVs were closely related, hybrid-type viruses with an ecotropic env gene and a xenotropic long terminal repeat. Duplications and a deletion were found in a restricted region of the hypervariable proline-rich region of Env glycoprotein.
Figures




Similar articles
-
Origins of the endogenous and infectious laboratory mouse gammaretroviruses.Viruses. 2014 Dec 26;7(1):1-26. doi: 10.3390/v7010001. Viruses. 2014. PMID: 25549291 Free PMC article. Review.
-
Acquisition of two endogenous ecotropic murine leukemia viruses in distinct Asian wild mouse populations.J Virol. 1991 Apr;65(4):1796-802. doi: 10.1128/JVI.65.4.1796-1802.1991. J Virol. 1991. PMID: 1672163 Free PMC article.
-
Precise identification of endogenous proviruses of NFS/N mice participating in recombination with moloney ecotropic murine leukemia virus (MuLV) to generate polytropic MuLVs.J Virol. 2005 Apr;79(8):4664-71. doi: 10.1128/JVI.79.8.4664-4671.2005. J Virol. 2005. PMID: 15795252 Free PMC article.
-
Diverse wild mouse origins of xenotropic, mink cell focus-forming, and two types of ecotropic proviral genes.J Virol. 1987 Oct;61(10):3082-8. doi: 10.1128/JVI.61.10.3082-3088.1987. J Virol. 1987. PMID: 3041030 Free PMC article.
-
Genetic control of retroviral disease in aging wild mice.Genetica. 1993;91(1-3):199-209. doi: 10.1007/BF01435998. Genetica. 1993. PMID: 8125269 Review.
Cited by
-
Origins of the endogenous and infectious laboratory mouse gammaretroviruses.Viruses. 2014 Dec 26;7(1):1-26. doi: 10.3390/v7010001. Viruses. 2014. PMID: 25549291 Free PMC article. Review.
-
Rmcf2, a xenotropic provirus in the Asian mouse species Mus castaneus, blocks infection by polytropic mouse gammaretroviruses.J Virol. 2005 Aug;79(15):9677-84. doi: 10.1128/JVI.79.15.9677-9684.2005. J Virol. 2005. PMID: 16014929 Free PMC article.
-
Exploring the role of endogenous retroviruses in seasonal reproductive cycles: a case study of the ERV-V envelope gene in mink.Front Cell Infect Microbiol. 2024 Jun 28;14:1404431. doi: 10.3389/fcimb.2024.1404431. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 39081866 Free PMC article.
-
Characterization of hortulanus endogenous murine leukemia virus, an endogenous provirus that encodes an infectious murine leukemia virus of a novel subgroup.J Virol. 2005 Jul;79(13):8316-29. doi: 10.1128/JVI.79.13.8316-8329.2005. J Virol. 2005. PMID: 15956577 Free PMC article.
-
Distribution of endogenous gammaretroviruses and variants of the Fv1 restriction gene in individual mouse strains and strain subgroups.PLoS One. 2019 Jul 10;14(7):e0219576. doi: 10.1371/journal.pone.0219576. eCollection 2019. PLoS One. 2019. PMID: 31291374 Free PMC article.
References
-
- Albritton L M, Tseng L, Scadden D, Cunningham J M. A putative murine ecotropic retrovirus receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell. 1989;57:659–666. - PubMed
-
- Bassin R H, Tuttle N, Fischinger P J. Rapid cell culture assay technic for murine leukaemia viruses. Nature. 1971;229:564–566. - PubMed
-
- Best S, Le Tissier P, Towers G, Stoye J P. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature. 1996;382:826–829. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

