The 3'-untranslated region of human interleukin-8 mRNA suppresses IL-8 gene expression
- PMID: 11328384
- PMCID: PMC1783198
- DOI: 10.1046/j.1365-2567.2001.01189.x
The 3'-untranslated region of human interleukin-8 mRNA suppresses IL-8 gene expression
Abstract
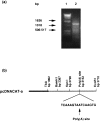
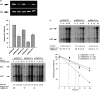
Although adenosine/uridine (AU)-rich sequences in the 3'-untranslated region (UTR) of the interleukin-8 (IL-8) gene have been suggested to contribute to its post-transcriptional regulation, the molecular basis whereby this occurs still needs to be understood. To investigate the role of the 3'-UTR on human IL-8 gene regulation, chimeric reporter genes were generated by adding full length or differentially deleted 3'-UTR of the IL-8 gene to chloramphenicol acetyltransferase (CAT). Addition of the entire IL-8 3'-UTR markedly reduced CAT mRNA and protein expression in COS 7 cells. In a reporter gene study, IL-8 3'-UTR destabilized CAT mRNA, which was dependent on active transcription in COS 7 cells. A 357-base sequence (nucleotides (nt) 2387-2743 of genomic DNA) within 3'-UTR, designated e, suppressed CAT gene expression by accelerating CAT mRNA turnover. A 26-base AU-rich sequence (nt 2552-2577) within e, containing four AUUUA pentamers that form two UAUUUAUU and one UUAUUUAU octamers, did not suppress CAT gene expression. However, deletion of the AU-rich sequences attenuated the inhibitory effect of e on CAT gene expression. Elimination of the first 100 bases (nt 2386-2486) attenuated the potency of fragment e, but much weaker than elimination of the first 146 bases (nt 2387-2533). This study gives new insights in unravelling the molecular mechanisms involved in the post-transcriptional regulation of the IL-8 gene.
Figures


Similar articles
-
The 3'-untranslated region of the mouse cholesterol 7alpha-hydroxylase mRNA contains elements responsive to post-transcriptional regulation by bile acids.Biochem J. 1997 Dec 1;328 ( Pt 2)(Pt 2):393-9. doi: 10.1042/bj3280393. Biochem J. 1997. PMID: 9371693 Free PMC article.
-
Regulation of an amino acid transporter mRNA in Trypanosoma brucei.Mol Biochem Parasitol. 2008 Jan;157(1):102-6. doi: 10.1016/j.molbiopara.2007.09.005. Epub 2007 Oct 2. Mol Biochem Parasitol. 2008. PMID: 17996963
-
The 3' untranslated region of IL-1beta regulates protein production.J Immunol. 1997 Feb 1;158(3):1187-93. J Immunol. 1997. PMID: 9013958
-
Posttranscriptional regulation of protein expression in human epithelial carcinoma cells by adenine-uridine-rich elements in the 3'-untranslated region of tumor necrosis factor-alpha messenger RNA.Cancer Res. 1997 Dec 1;57(23):5426-33. Cancer Res. 1997. PMID: 9393771
-
Functionally independent AU-rich sequence motifs regulate KC (CXCL1) mRNA.J Biol Chem. 2005 Aug 26;280(34):30166-74. doi: 10.1074/jbc.M502280200. Epub 2005 Jul 1. J Biol Chem. 2005. PMID: 15994316
Cited by
-
IL-17 attenuates degradation of ARE-mRNAs by changing the cooperation between AU-binding proteins and microRNA16.PLoS Genet. 2013;9(9):e1003747. doi: 10.1371/journal.pgen.1003747. Epub 2013 Sep 26. PLoS Genet. 2013. PMID: 24086143 Free PMC article.
-
Distinct domains of AU-rich elements exert different functions in mRNA destabilization and stabilization by p38 mitogen-activated protein kinase or HuR.Mol Cell Biol. 2004 Jun;24(11):4835-47. doi: 10.1128/MCB.24.11.4835-4847.2004. Mol Cell Biol. 2004. PMID: 15143177 Free PMC article.
-
The Formulation of the N-Acetylglucosamine as Nanoparticles Increases Its Anti-Inflammatory Activities: An In Vitro Study.Bioengineering (Basel). 2023 Mar 9;10(3):343. doi: 10.3390/bioengineering10030343. Bioengineering (Basel). 2023. PMID: 36978734 Free PMC article.
-
Novel AU-rich proximal UTR sequences (APS) enhance CXCL8 synthesis upon the induction of rpS6 phosphorylation.PLoS Genet. 2019 Apr 10;15(4):e1008077. doi: 10.1371/journal.pgen.1008077. eCollection 2019 Apr. PLoS Genet. 2019. PMID: 30969964 Free PMC article.
-
Tip110/SART3 regulates IL-8 expression and predicts the clinical outcomes in melanoma.Mol Cancer. 2018 Aug 17;17(1):124. doi: 10.1186/s12943-018-0868-z. Mol Cancer. 2018. PMID: 30119675 Free PMC article.
References
-
- Baggiolini M, Dewald B, Moser B. Interleukin-8 and related chemotactic cytokines-CXC and CC chemokines. Adv Immunol. 1994;55:97–197. - PubMed
-
- Baggiolini M, Dewald B, Moser B. Human chemokines: an update. Annu Rev Immunol. 1997;15:675–705. - PubMed
-
- Gillitzer R, Berger R, Meilke V, Miler C, Wolff K, Sting G. Upper keratinocytes of psoriatic skin lesions express high levels of NAP-1/IL-8 mRNA in situ. J Invest Dermatol. 1991;97:73–9. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

