Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations
- PMID: 11312331
- PMCID: PMC114214
- DOI: 10.1128/JVI.75.10.4614-4624.2001
Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations
Abstract
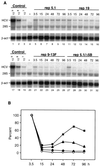
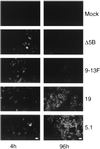
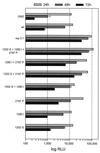
Studies of the Hepatitis C virus (HCV) replication cycle have been made possible with the development of subgenomic selectable RNAs that replicate autonomously in cultured cells. In these replicons the region encoding the HCV structural proteins was replaced by the neomycin phosphotransferase gene, allowing the selection of transfected cells that support high-level replication of these RNAs. Subsequent analyses revealed that, within selected cells, HCV RNAs had acquired adaptive mutations that increased the efficiency of colony formation by an unknown mechanism. Using a panel of replicons that differed in their degrees of cell culture adaptation, in this study we show that adaptive mutations enhance RNA replication. Transient-transfection assays that did not require selection of transfected cells demonstrated a clear correlation between the level of adaptation and RNA replication. The highest replication level was found with an adapted replicon carrying two amino acid substitutions located in NS3 and one in NS5A that acted synergistically. In contrast, the nonadapted RNA replicated only transiently and at a low level. The correlation between the efficiency of colony formation and RNA replication was corroborated with replicons in which the selectable marker gene was replaced by the gene encoding firefly luciferase. Upon transfection of naive Huh-7 cells, the levels of luciferase activity directly reflected the replication efficiencies of the various replicon RNAs. These results show that cell culture-adaptive mutations enhance HCV RNA replication.
Figures


Similar articles
-
Dominant negative effect of wild-type NS5A on NS5A-adapted subgenomic hepatitis C virus RNA replicon.J Gen Virol. 2004 Jul;85(Pt 7):1867-1875. doi: 10.1099/vir.0.80006-0. J Gen Virol. 2004. PMID: 15218171
-
Trans-complementation of HCV replication by non-structural protein 5A.Virus Res. 2006 Feb;115(2):122-30. doi: 10.1016/j.virusres.2005.07.012. Epub 2005 Sep 15. Virus Res. 2006. PMID: 16146661
-
Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees.Proc Natl Acad Sci U S A. 2002 Oct 29;99(22):14416-21. doi: 10.1073/pnas.212532699. Epub 2002 Oct 21. Proc Natl Acad Sci U S A. 2002. PMID: 12391335 Free PMC article.
-
[Replication of hepatitis C virus genome].Uirusu. 2008 Dec;58(2):191-8. Uirusu. 2008. PMID: 19374197 Review. Japanese.
-
[Interferon resistance and ISDR (interferon sensitivity determining region)].Nihon Rinsho. 2006 Jul;64(7):1249-53. Nihon Rinsho. 2006. PMID: 16838640 Review. Japanese.
Cited by
-
Relation between viral fitness and immune escape within the hepatitis C virus protease.Gut. 2006 Feb;55(2):266-74. doi: 10.1136/gut.2005.072231. Epub 2005 Aug 16. Gut. 2006. PMID: 16105887 Free PMC article.
-
Mutagenesis analysis of the rGTP-specific binding site of hepatitis C virus RNA-dependent RNA polymerase.J Virol. 2005 Sep;79(18):11607-17. doi: 10.1128/JVI.79.18.11607-11617.2005. J Virol. 2005. PMID: 16140738 Free PMC article.
-
Replication fitness and NS5B drug sensitivity of diverse hepatitis C virus isolates characterized by using a transient replication assay.Antimicrob Agents Chemother. 2005 May;49(5):2059-69. doi: 10.1128/AAC.49.5.2059-2069.2005. Antimicrob Agents Chemother. 2005. PMID: 15855532 Free PMC article.
-
Structural and functional analysis of hepatitis C virus strain JFH1 polymerase.J Virol. 2009 Nov;83(22):11926-39. doi: 10.1128/JVI.01008-09. Epub 2009 Sep 9. J Virol. 2009. PMID: 19740982 Free PMC article.
-
Interferon regulatory factor 3-independent double-stranded RNA-induced inhibition of hepatitis C virus replicons in human embryonic kidney 293 cells.J Virol. 2005 Mar;79(5):3174-8. doi: 10.1128/JVI.79.5.3174-3178.2005. J Virol. 2005. PMID: 15709037 Free PMC article.
References
-
- Bartenschlager R, Lohmann V. Replication of hepatitis C virus. J Gen Virol. 2000;81:1631–1648. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

