Regulation and localization of the Bloom syndrome protein in response to DNA damage
- PMID: 11309417
- PMCID: PMC2169463
- DOI: 10.1083/jcb.153.2.367
Regulation and localization of the Bloom syndrome protein in response to DNA damage
Abstract
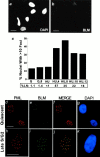
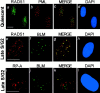
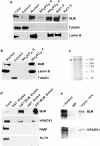
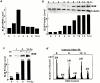
Bloom syndrome (BS) is an autosomal recessive disorder characterized by a high incidence of cancer and genomic instability. BLM, the protein defective in BS, is a RecQ-like helicase, presumed to function in DNA replication, recombination, or repair. BLM localizes to promyelocytic leukemia protein (PML) nuclear bodies and is expressed during late S and G2. We show, in normal human cells, that the recombination/repair proteins hRAD51 and replication protein (RP)-A assembled with BLM into a fraction of PML bodies during late S/G2. Biochemical experiments suggested that BLM resides in a nuclear matrix-bound complex in which association with hRAD51 may be direct. DNA-damaging agents that cause double strand breaks and a G2 delay induced BLM by a p53- and ataxia-telangiectasia mutated independent mechanism. This induction depended on the G2 delay, because it failed to occur when G2 was prevented or bypassed. It coincided with the appearance of foci containing BLM, PML, hRAD51 and RP-A, which resembled ionizing radiation-induced foci. After radiation, foci containing BLM and PML formed at sites of single-stranded DNA and presumptive repair in normal cells, but not in cells with defective PML. Our findings suggest that BLM is part of a dynamic nuclear matrix-based complex that requires PML and functions during G2 in undamaged cells and recombinational repair after DNA damage.
Figures
Similar articles
-
Phosphorylation of BLM, dissociation from topoisomerase IIIalpha, and colocalization with gamma-H2AX after topoisomerase I-induced replication damage.Mol Cell Biol. 2005 Oct;25(20):8925-37. doi: 10.1128/MCB.25.20.8925-8937.2005. Mol Cell Biol. 2005. PMID: 16199871 Free PMC article.
-
A role for PML and the nuclear body in genomic stability.Oncogene. 1999 Dec 23;18(56):7941-7. doi: 10.1038/sj.onc.1203367. Oncogene. 1999. PMID: 10637504
-
BLM helicase-dependent transport of p53 to sites of stalled DNA replication forks modulates homologous recombination.EMBO J. 2003 Mar 3;22(5):1210-22. doi: 10.1093/emboj/cdg114. EMBO J. 2003. PMID: 12606585 Free PMC article.
-
Clinical features of Bloom syndrome and function of the causative gene, BLM helicase.Expert Rev Mol Diagn. 2004 May;4(3):393-401. doi: 10.1586/14737159.4.3.393. Expert Rev Mol Diagn. 2004. PMID: 15137905 Review.
-
Functions of RecQ family helicases: possible involvement of Bloom's and Werner's syndrome gene products in guarding genome integrity during DNA replication.J Biochem. 2001 Apr;129(4):501-7. doi: 10.1093/oxfordjournals.jbchem.a002883. J Biochem. 2001. PMID: 11275547 Review.
Cited by
-
A TRilogy of ATR's Non-Canonical Roles Throughout the Cell Cycle and Its Relation to Cancer.Cancers (Basel). 2024 Oct 19;16(20):3536. doi: 10.3390/cancers16203536. Cancers (Basel). 2024. PMID: 39456630 Free PMC article. Review.
-
PML Nuclear bodies: the cancer connection and beyond.Nucleus. 2024 Dec;15(1):2321265. doi: 10.1080/19491034.2024.2321265. Epub 2024 Feb 27. Nucleus. 2024. PMID: 38411156 Free PMC article. Review.
-
BLAP75, an essential component of Bloom's syndrome protein complexes that maintain genome integrity.EMBO J. 2005 Apr 6;24(7):1465-76. doi: 10.1038/sj.emboj.7600622. Epub 2005 Mar 17. EMBO J. 2005. PMID: 15775963 Free PMC article.
-
Importance of Promyelocytic Leukema Protein (PML) for Kaposi's Sarcoma-Associated Herpesvirus Lytic Replication.Front Microbiol. 2018 Oct 8;9:2324. doi: 10.3389/fmicb.2018.02324. eCollection 2018. Front Microbiol. 2018. PMID: 30349510 Free PMC article.
-
BLM Deficiency Is Not Associated with Sensitivity to Hydroxyurea-Induced Replication Stress.J Nucleic Acids. 2010 Sep 8;2010:319754. doi: 10.4061/2010/319754. J Nucleic Acids. 2010. PMID: 20936166 Free PMC article.
References
-
- Aurias A., Antoine J.L., Assathiany R., Odievre M., Dutrillaux B. Radiation sensitivity of Bloom's syndrome lymphocytes during S and G2 phases. Cancer Genet. Cytogenet. 1985;16:131–136. - PubMed
-
- Banin S., Moyal L., Shieh S., Taya Y., Anderson C.W., Chessa L., Smorodinsky N.I., Prives C., Reiss Y., Shiloh Y., Ziv Y. Enhanced phosphorylation of p53 by ATM in response to DNA damage. Science. 1998;281:1674–1677. - PubMed
-
- Beamish H., Khanna K.K., Lavin M.F. Ionizing radiation and cell cycle progression in ataxia telangiectasia. Radiat. Res. 1994;138:130–133. - PubMed
-
- Brosh R.M., Li J.L., Kenny M.K., Karow J.K., Cooper M.P., Kureekattil R.P., Hickson I.D., Bohr V.A. Replication protein A physically interacts with the Bloom's syndrome protein and stimulates its helicase activity. J. Biol. Chem. 2000;275:23500–23508. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

