Glucocorticoid-induced DNA demethylation and gene memory during development
- PMID: 11296230
- PMCID: PMC125428
- DOI: 10.1093/emboj/20.8.1974
Glucocorticoid-induced DNA demethylation and gene memory during development
Abstract
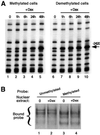
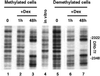
Glucocorticoid hormones were found to regulate DNA demethylation within a key enhancer of the rat liver-specific tyrosine aminotransferase (Tat) gene. Genomic footprinting analysis shows that the glucocorticoid receptor uses local DNA demethylation as one of several steps to recruit transcription factors in hepatoma cells. Demethylation occurs within 2-3 days following rapid (< 1 h) chromatin remodeling and recruitment of a first transcription factor, HNF-3. Upon demethylation, two additional transcription factors are recruited when chromatin is remodeled. In contrast to chromatin remodeling, the demethylation is stable following hormone withdrawal. As a stronger subsequent glucocorticoid response is observed, demethylation appears to provide memory of the first stimulation. During development, this demethylation occurs before birth, at a stage where the Tat gene is not yet inducible, and it could thus prepare the enhancer for subsequent stimulation by hypoglycemia at birth. In vitro cultures of fetal hepatocytes recapitulate the regulation analyzed in hepatoma cells. There fore, demethylation appears to contribute to the fine-tuning of the enhancer and to the memorization of a regulatory event during development.
Figures

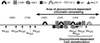


Similar articles
-
In vivo analysis of the model tyrosine aminotransferase gene reveals multiple sequential steps in glucocorticoid receptor action.Oncogene. 2001 May 28;20(24):3028-38. doi: 10.1038/sj.onc.1204327. Oncogene. 2001. PMID: 11420718 Review.
-
Hepatocyte-specific interplay of transcription factors at the far-upstream enhancer of the carbamoylphosphate synthetase gene upon glucocorticoid induction.FEBS J. 2007 Jan;274(1):37-45. doi: 10.1111/j.1742-4658.2006.05561.x. Epub 2006 Nov 28. FEBS J. 2007. PMID: 17140418
-
Nature of the accessible chromatin at a glucocorticoid-responsive enhancer.Mol Cell Biol. 2004 Sep;24(18):7891-901. doi: 10.1128/MCB.24.18.7891-7901.2004. Mol Cell Biol. 2004. PMID: 15340052 Free PMC article.
-
Divergent regulation of the class II P-glycoprotein gene in primary cultures of hepatocytes versus H35 hepatoma by glucocorticoids.Cell Growth Differ. 1995 Oct;6(10):1321-32. Cell Growth Differ. 1995. PMID: 8845310
-
Local DNA demethylation in vertebrates: how could it be performed and targeted?FEBS Lett. 2001 Apr 13;494(3):135-40. doi: 10.1016/s0014-5793(01)02328-6. FEBS Lett. 2001. PMID: 11311228 Review.
Cited by
-
In utero exposure to cigarette chemicals induces sex-specific disruption of one-carbon metabolism and DNA methylation in the human fetal liver.BMC Med. 2015 Jan 29;13:18. doi: 10.1186/s12916-014-0251-x. BMC Med. 2015. PMID: 25630355 Free PMC article.
-
Impact of oxidative stress in fetal programming.J Pregnancy. 2012;2012:582748. doi: 10.1155/2012/582748. Epub 2012 Jul 11. J Pregnancy. 2012. PMID: 22848830 Free PMC article. Review.
-
The Signature of Maternal Social Rank in Placenta Deoxyribonucleic Acid Methylation Profiles in Rhesus Monkeys.Child Dev. 2017 May;88(3):900-918. doi: 10.1111/cdev.12640. Epub 2016 Oct 14. Child Dev. 2017. PMID: 27739069 Free PMC article.
-
Retinoic acid receptor-related orphan receptor (ROR) alpha4 is the predominant isoform of the nuclear receptor RORalpha in the liver and is up-regulated by hypoxia in HepG2 human hepatoma cells.Biochem J. 2002 Jun 1;364(Pt 2):449-56. doi: 10.1042/BJ20011558. Biochem J. 2002. PMID: 12023888 Free PMC article.
-
Identification of dynamic glucocorticoid-induced methylation changes at the FKBP5 locus.Clin Epigenetics. 2019 May 23;11(1):83. doi: 10.1186/s13148-019-0682-5. Clin Epigenetics. 2019. PMID: 31122292 Free PMC article.
References
-
- Antequera F.J., Boyes,J. and Bird,A. (1990) High levels of de novo methylation and altered chromatin structure at CpG islands in cell lines. Cell, 6, 503–514. - PubMed
-
- Bird A.P. (1995) Gene number, noise reduction and biological complexity. Trends Genet., 11, 94–100. - PubMed
-
- Bird A.P. and Wolffe,A.P. (1999) Methylation-induced repression—belts, braces, and chromatin. Cell, 99, 451–454. - PubMed
-
- Boyes J. and Bird,A. (1991) DNA methylation inhibits transcription indirectly via a methyl-CpG binding protein. Cell, 64, 1123–1134. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

