High-mobility-group proteins NHP6A and NHP6B participate in activation of the RNA polymerase III SNR6 gene
- PMID: 11287614
- PMCID: PMC86937
- DOI: 10.1128/MCB.21.9.3096-3104.2001
High-mobility-group proteins NHP6A and NHP6B participate in activation of the RNA polymerase III SNR6 gene
Abstract
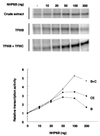
Transcription of yeast class III genes involves the formation of a transcription initiation complex that comprises RNA polymerase III (Pol III) and the general transcription factors TFIIIB and TFIIIC. Using a genetic screen for positive regulators able to compensate for a deficiency in a promoter element of the SNR6 gene, we isolated the NHP6A and NHP6B genes. Here we show that the high-mobility-group proteins NHP6A and NHP6B are required for the efficient transcription of the SNR6 gene both in vivo and in vitro. The transcripts of wild-type and promoter-defective SNR6 genes decreased or became undetectable in an nhp6ADelta nhp6BDelta double-mutant strain, and the protection over the TATA box of the wild-type SNR6 gene was lost in nhp6ADelta nhp6BDelta cells at 37 degrees C. In vitro, NHP6B specifically stimulated the transcription of SNR6 templates up to fivefold in transcription assays using either cell nuclear extracts from nhp6ADelta nhp6BDelta cells or reconstituted transcription systems. Finally, NHP6B activated SNR6 transcription in a TFIIIC-independent assay. These results indicate that besides the general transcription factors TFIIIB and TFIIIC, additional auxillary factors are required for the optimal transcription of at least some specific Pol III genes.
Figures


Similar articles
-
Nhp6, an HMG1 protein, functions in SNR6 transcription by RNA polymerase III in S. cerevisiae.Mol Cell. 2001 Feb;7(2):309-18. doi: 10.1016/s1097-2765(01)00179-4. Mol Cell. 2001. PMID: 11239460
-
A novel upstream RNA polymerase III promoter element becomes essential when the chromatin structure of the yeast U6 RNA gene is altered.Mol Cell Biol. 2001 Oct;21(19):6429-39. doi: 10.1128/MCB.21.19.6429-6439.2001. Mol Cell Biol. 2001. PMID: 11533232 Free PMC article.
-
Identical components of yeast transcription factor IIIB are required and sufficient for transcription of TATA box-containing and TATA-less genes.Mol Cell Biol. 1994 Apr;14(4):2798-808. doi: 10.1128/mcb.14.4.2798-2808.1994. Mol Cell Biol. 1994. PMID: 8139577 Free PMC article.
-
Extra-transcriptional functions of RNA Polymerase III complexes: TFIIIC as a potential global chromatin bookmark.Gene. 2012 Feb 10;493(2):169-75. doi: 10.1016/j.gene.2011.09.018. Epub 2011 Oct 1. Gene. 2012. PMID: 21986035 Review.
-
Gene insulation. Part I: natural strategies in yeast and Drosophila.Biochem Cell Biol. 2010 Dec;88(6):875-84. doi: 10.1139/O10-110. Biochem Cell Biol. 2010. PMID: 21102650 Review.
Cited by
-
Regulation of pol III transcription by nutrient and stress signaling pathways.Biochim Biophys Acta. 2013 Mar-Apr;1829(3-4):361-75. doi: 10.1016/j.bbagrm.2012.11.001. Epub 2012 Nov 16. Biochim Biophys Acta. 2013. PMID: 23165150 Free PMC article. Review.
-
Reconfiguring the connectivity of a multiprotein complex: fusions of yeast TATA-binding protein with Brf1, and the function of transcription factor IIIB.Proc Natl Acad Sci U S A. 2005 Oct 25;102(43):15406-11. doi: 10.1073/pnas.0507653102. Epub 2005 Oct 14. Proc Natl Acad Sci U S A. 2005. PMID: 16227432 Free PMC article.
-
Nhp6: a small but powerful effector of chromatin structure in Saccharomyces cerevisiae.Biochim Biophys Acta. 2010 Jan-Feb;1799(1-2):175-80. doi: 10.1016/j.bbagrm.2009.11.010. Biochim Biophys Acta. 2010. PMID: 20123079 Free PMC article. Review.
-
MRN1 implicates chromatin remodeling complexes and architectural factors in mRNA maturation.PLoS One. 2012;7(9):e44373. doi: 10.1371/journal.pone.0044373. Epub 2012 Sep 18. PLoS One. 2012. PMID: 23028530 Free PMC article.
-
Position-dependent function of a B block promoter element implies a specialized chromatin structure on the S.cerevisiae U6 RNA gene, SNR6.Nucleic Acids Res. 2004 Aug 10;32(14):4297-305. doi: 10.1093/nar/gkh769. Print 2004. Nucleic Acids Res. 2004. PMID: 15304565 Free PMC article.
References
-
- Aasland R, Stewart A F, Gibson T. The SANT domain: a putative DNA-binding domain in the SWI-SNF and ADA complexes, the transcriptional co-repressor N-CoR and TFIIIB. Trends Biochem Sci. 1996;21:87–88. - PubMed
-
- Brow D A, Guthrie C. Transcription of a yeast U6 snRNA gene requires a polymerase III promoter element in a novel position. Genes Dev. 1990;4:1345–1356. - PubMed
-
- Buratowski S, Zhou H. A suppressor of TBP mutations encodes an RNA polymerase III transcription factor with homology to TFIIB. Cell. 1992;71:221–230. - PubMed
-
- Burnol A F, Margottin F, Schultz P, Marsolier M C, Oudet P, Sentenac A. Basal promoter and enhancer element of yeast U6 snRNA gene. J Mol Biol. 1993;233:644–658. - PubMed
-
- Bustin M, Reeves R. High-mobility-group chromosomal proteins: architectural components that facilitate chromatin function. Prog Nucleic Acid Res Mol Biol. 1996;54:35–100. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
