Mutational evidence for an internal fusion peptide in flavivirus envelope protein E
- PMID: 11287576
- PMCID: PMC114172
- DOI: 10.1128/JVI.75.9.4268-4275.2001
Mutational evidence for an internal fusion peptide in flavivirus envelope protein E
Abstract
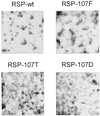
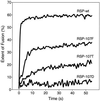
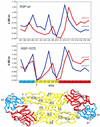
The envelope protein E of the flavivirus tick-borne encephalitis (TBE) virus promotes cell entry by inducing fusion of the viral membrane with an intracellular membrane after uptake by endocytosis. This protein differs from other well-studied viral and cellular fusion proteins because of its distinct molecular architecture and apparent lack of involvement of coiled coils in the low-pH-induced structural transitions that lead to fusion. A highly conserved loop (the cd loop), which resides at the distal tip of each subunit and is mostly buried in the subunit interface of the native E homodimer at neutral pH, has been hypothesized to function as an internal fusion peptide at low pH, but this has not yet been shown experimentally. It was predicted by examination of the X-ray crystal structure of the TBE virus E protein (F. A. Rey et al., Nature 375:291-298, 1995) that mutations at a specific residue within this loop (Leu 107) would not cause the native structure to be disrupted. We therefore introduced amino acid substitutions at this position and, using recombinant subviral particles, investigated the effects of these changes on fusion and related properties. Replacement of Leu with hydrophilic amino acids strongly impaired (Thr) or abolished (Asp) fusion activity, whereas a Phe mutant still retained a significant degree of fusion activity. Liposome coflotation experiments showed that the fusion-negative Asp mutant did not form a stable interaction with membranes at low pH, although it was still capable of undergoing the structural rearrangements required for fusion. These data support the hypothesis that the cd loop may be directly involved in interactions with target membranes during fusion.
Figures




Similar articles
-
Membrane fusion activity of tick-borne encephalitis virus and recombinant subviral particles in a liposomal model system.Virology. 2000 Mar 30;269(1):37-46. doi: 10.1006/viro.1999.0172. Virology. 2000. PMID: 10725196
-
Membrane interactions of the tick-borne encephalitis virus fusion protein E at low pH.J Virol. 2002 Apr;76(8):3784-90. doi: 10.1128/jvi.76.8.3784-3790.2002. J Virol. 2002. PMID: 11907218 Free PMC article.
-
Identification of specific histidines as pH sensors in flavivirus membrane fusion.J Cell Biol. 2008 Oct 20;183(2):353-61. doi: 10.1083/jcb.200806081. J Cell Biol. 2008. PMID: 18936253 Free PMC article.
-
Molecular mechanisms of flavivirus membrane fusion.Amino Acids. 2011 Nov;41(5):1159-63. doi: 10.1007/s00726-009-0370-4. Epub 2009 Nov 1. Amino Acids. 2011. PMID: 19882217 Review.
-
The machinery for flavivirus fusion with host cell membranes.Curr Opin Microbiol. 2001 Aug;4(4):450-5. doi: 10.1016/s1369-5274(00)00234-4. Curr Opin Microbiol. 2001. PMID: 11495810 Review.
Cited by
-
Small-molecule inhibitors of dengue-virus entry.PLoS Pathog. 2012;8(4):e1002627. doi: 10.1371/journal.ppat.1002627. Epub 2012 Apr 5. PLoS Pathog. 2012. PMID: 22496653 Free PMC article.
-
Hydrophobic residues that form putative fusion loops of Epstein-Barr virus glycoprotein B are critical for fusion activity.J Virol. 2007 Sep;81(17):9596-600. doi: 10.1128/JVI.00758-07. Epub 2007 Jun 6. J Virol. 2007. PMID: 17553877 Free PMC article.
-
Targeted delivery of small interfering RNA to human dendritic cells to suppress dengue virus infection and associated proinflammatory cytokine production.J Virol. 2010 Mar;84(5):2490-501. doi: 10.1128/JVI.02105-08. Epub 2009 Dec 16. J Virol. 2010. PMID: 20015996 Free PMC article.
-
Structures of immature flavivirus particles.EMBO J. 2003 Jun 2;22(11):2604-13. doi: 10.1093/emboj/cdg270. EMBO J. 2003. PMID: 12773377 Free PMC article.
-
The stoichiometry of antibody-mediated neutralization and enhancement of West Nile virus infection.Cell Host Microbe. 2007 Apr 19;1(2):135-45. doi: 10.1016/j.chom.2007.03.002. Cell Host Microbe. 2007. PMID: 18005691 Free PMC article.
References
-
- Allison S L, Mandl C W, Kunz C, Heinz F X. Expression of cloned envelope protein genes from the flavivirus tick-borne encephalitis virus in mammalian cells and random mutagenesis by PCR. Virus Genes. 1994;8:187–198. - PubMed
-
- Baker K A, Dutch R E, Lamb R A, Jardetzky T S. Structural basis for paramyxovirus-mediated membrane fusion. Mol Cell. 1999;3:309–319. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

