An essential role for the substrate-binding region of Hsp40s in Saccharomyces cerevisiae
- PMID: 11266475
- PMCID: PMC2195774
- DOI: 10.1083/jcb.152.4.851
An essential role for the substrate-binding region of Hsp40s in Saccharomyces cerevisiae
Abstract
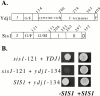
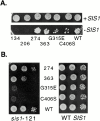
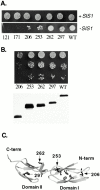
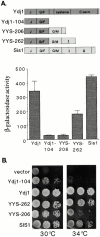
In addition to regulating the ATPase cycle of Hsp70, a second critical role of Hsp40s has been proposed based on in vitro studies: binding to denatured protein substrates, followed by their presentation to Hsp70 for folding. However, the biological importance of this model is challenged by the fact that deletion of the substrate-binding domain of either of the two major Hsp40s of the yeast cytosol, Ydj1 and Sis1, leads to no severe defects, as long as regions necessary for Hsp70 interaction are retained. As an in vivo test of this model, requirements for viability were examined in a strain having deletions of both Hsp40 genes. Despite limited sequence similarity, the substrate-binding domain of either Sis1 or Ydj1 allowed cell growth, indicating they share overlapping essential functions. Furthermore, the substrate-binding domain must function in cis with a functional Hsp70-interacting domain. We conclude that the ability of cytosolic Hsp40s to bind unfolded protein substrates is an essential function in vivo.
Figures

Similar articles
-
The glycine-phenylalanine-rich region determines the specificity of the yeast Hsp40 Sis1.Mol Cell Biol. 1999 Nov;19(11):7751-8. doi: 10.1128/MCB.19.11.7751. Mol Cell Biol. 1999. PMID: 10523664 Free PMC article.
-
Exchangeable chaperone modules contribute to specification of type I and type II Hsp40 cellular function.Mol Biol Cell. 2004 Feb;15(2):761-73. doi: 10.1091/mbc.e03-03-0146. Epub 2003 Dec 2. Mol Biol Cell. 2004. PMID: 14657253 Free PMC article.
-
Class-specific interactions between Sis1 J-domain protein and Hsp70 chaperone potentiate disaggregation of misfolded proteins.Proc Natl Acad Sci U S A. 2021 Dec 7;118(49):e2108163118. doi: 10.1073/pnas.2108163118. Proc Natl Acad Sci U S A. 2021. PMID: 34873058 Free PMC article.
-
Role of J-domain Proteins in Yeast Physiology and Protein Quality Control.J Mol Biol. 2024 Jul 15;436(14):168484. doi: 10.1016/j.jmb.2024.168484. Epub 2024 Feb 7. J Mol Biol. 2024. PMID: 38331212 Review.
-
Specification of Hsp70 function by Type I and Type II Hsp40.Subcell Biochem. 2015;78:91-102. doi: 10.1007/978-3-319-11731-7_4. Subcell Biochem. 2015. PMID: 25487017 Review.
Cited by
-
Swa2, the yeast homolog of mammalian auxilin, is specifically required for the propagation of the prion variant [URE3-1].Mol Microbiol. 2015 Sep;97(5):926-41. doi: 10.1111/mmi.13076. Epub 2015 Jun 25. Mol Microbiol. 2015. PMID: 26031938 Free PMC article.
-
J Proteins Counteract Amyloid Propagation and Toxicity in Yeast.Biology (Basel). 2022 Aug 30;11(9):1292. doi: 10.3390/biology11091292. Biology (Basel). 2022. PMID: 36138771 Free PMC article. Review.
-
Mechanisms of the Hsp70 chaperone system.Biochem Cell Biol. 2010 Apr;88(2):291-300. doi: 10.1139/o09-175. Biochem Cell Biol. 2010. PMID: 20453930 Free PMC article. Review.
-
The Role of HSP40 Conserved Motifs in the Response to Cytotoxic Stress.J Nat Sci. 2018;4(4):e500. J Nat Sci. 2018. PMID: 29682607 Free PMC article.
-
The Genomes of Three Uneven Siblings: Footprints of the Lifestyles of Three Trichoderma Species.Microbiol Mol Biol Rev. 2016 Feb 10;80(1):205-327. doi: 10.1128/MMBR.00040-15. Print 2016 Mar. Microbiol Mol Biol Rev. 2016. PMID: 26864432 Free PMC article. Review.
References
-
- Banecki B., Liberek K., Wall D., Wawrzynow A., Georgopoulos C., Bertoli E., Tanfani F., Zylicz M. Structure-function analysis of the zinc finger region of the DnaJ molecular chaperone. J. Biol. Chem. 1996;271:14840–14848. - PubMed
-
- Bukau B., Horwich A.L. The Hsp70 and Hsp60 chaperone machines. Cell. 1998;92:351–366. - PubMed
-
- Caplan A.J., Cyr D.M., Douglas M.G. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism Cell. 71 1992. 1143 1155a - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

