CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs
- PMID: 11226150
- PMCID: PMC140195
- DOI: 10.1093/emboj/20.1.12
CD81 extracellular domain 3D structure: insight into the tetraspanin superfamily structural motifs
Abstract
Human CD81, a known receptor for hepatitis C virus envelope E2 glycoprotein, is a transmembrane protein belonging to the tetraspanin family. The crystal structure of human CD81 large extracellular domain is reported here at 1.6 A resolution. Each subunit within the homodimeric protein displays a mushroom-like structure, composed of five alpha-helices arranged in 'stalk' and 'head' subdomains. Residues known to be involved in virus binding can be mapped onto the head subdomain, providing a basis for the design of antiviral drugs and vaccines. Sequence analysis of 160 tetraspanins indicates that key structural features and the new protein fold observed in the CD81 large extracellular domain are conserved within the family. On these bases, it is proposed that tetraspanins may assemble at the cell surface into homo- and/or hetero-dimers through a conserved hydrophobic interface located in the stalk subdomain, while interacting with other liganding proteins, including hepatitis C virus E2, through the head subdomain. The topology of such interactions provides a rationale for the assembly of the so-called tetraspan-web.
Figures

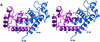
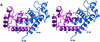
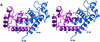

Similar articles
-
[Structural biology of human CD81, a receptor for hepatitis C virus].Uirusu. 2004 Jun;54(1):39-47. doi: 10.2222/jsv.54.39. Uirusu. 2004. PMID: 15449903 Review. Japanese.
-
Crystallization and preliminary crystallographic studies on the large extracellular domain of human CD81, a tetraspanin receptor for hepatitis C virus.Acta Crystallogr D Biol Crystallogr. 2001 Jan;57(Pt 1):156-8. doi: 10.1107/s0907444900015468. Acta Crystallogr D Biol Crystallogr. 2001. PMID: 11134943
-
Subunit association and conformational flexibility in the head subdomain of human CD81 large extracellular loop.Biol Chem. 2002 Sep;383(9):1447-52. doi: 10.1515/BC.2002.164. Biol Chem. 2002. PMID: 12437138
-
Complete predicted three-dimensional structure of the facilitator transmembrane protein and hepatitis C virus receptor CD81: conserved and variable structural domains in the tetraspanin superfamily.Biophys J. 2006 Jan 1;90(1):212-27. doi: 10.1529/biophysj.105.069666. Biophys J. 2006. PMID: 16352525 Free PMC article.
-
Structural characterization of CD81-Claudin-1 hepatitis C virus receptor complexes.Biochem Soc Trans. 2011 Apr;39(2):537-40. doi: 10.1042/BST0390537. Biochem Soc Trans. 2011. PMID: 21428935 Review.
Cited by
-
Oligomerization of the Tetraspanin CD81 via the Flexibility of Its δ-Loop.Biophys J. 2016 Jun 7;110(11):2463-2474. doi: 10.1016/j.bpj.2016.05.003. Biophys J. 2016. PMID: 27276264 Free PMC article.
-
Ultrasonic imaging of endothelial CD81 expression using CD81-targeted contrast agents in in vitro and in vivo studies.Ultrasound Med Biol. 2012 Apr;38(4):670-80. doi: 10.1016/j.ultrasmedbio.2011.12.025. Epub 2012 Feb 16. Ultrasound Med Biol. 2012. PMID: 22341598 Free PMC article.
-
Hepatitis C Virus Structure: Defined by What It Is Not.Cold Spring Harb Perspect Med. 2020 Jan 2;10(1):a036822. doi: 10.1101/cshperspect.a036822. Cold Spring Harb Perspect Med. 2020. PMID: 31501263 Free PMC article. Review.
-
Interaction of hepatitis C virus envelope glycoprotein E2 with the large extracellular loop of tupaia CD81.World J Gastroenterol. 2009 Jan 14;15(2):240-4. doi: 10.3748/wjg.15.240. World J Gastroenterol. 2009. PMID: 19132776 Free PMC article.
-
Targeting host factors: a novel rationale for the management of hepatitis C virus.World J Gastroenterol. 2009 Jul 28;15(28):3472-9. doi: 10.3748/wjg.15.3472. World J Gastroenterol. 2009. PMID: 19630100 Free PMC article. Review.
References
-
- Abrahams J.P. and Leslie,A.G.W. (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D, 52, 30–42. - PubMed
-
- Brünger A.T. et al. (1998) Crystallography & NMR system (CNS): a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. - PubMed
-
- Cha J.H., Brooke,J.S., Ivey,K.N. and Eidels,L. (2000) Cell surface monkey CD9 antigen is a coreceptor that increases diphtheria toxin sensitivity and diphtheria toxin receptor affinity. J. Biol. Chem., 275, 6901–6907. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

