Lytic replication of Kaposi's sarcoma-associated herpesvirus results in the formation of multiple capsid species: isolation and molecular characterization of A, B, and C capsids from a gammaherpesvirus
- PMID: 11222712
- PMCID: PMC115913
- DOI: 10.1128/JVI.75.6.2866-2878.2001
Lytic replication of Kaposi's sarcoma-associated herpesvirus results in the formation of multiple capsid species: isolation and molecular characterization of A, B, and C capsids from a gammaherpesvirus
Abstract
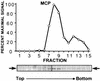
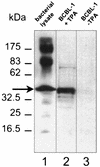
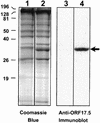
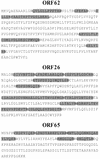
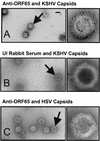
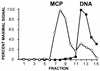
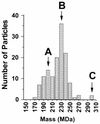
Despite the discovery of Epstein-Barr virus more than 35 years ago, a thorough understanding of gammaherpesvirus capsid composition and structure has remained elusive. We approached this problem by purifying capsids from Kaposi's sarcoma-associated herpesvirus (KSHV), the only other known human gammaherpesvirus. The results from our biochemical and imaging analyses demonstrate that KSHV capsids possess a typical herpesvirus icosahedral capsid shell composed of four structural proteins. The hexameric and pentameric capsomers are composed of the major capsid protein (MCP) encoded by open reading frame 25. The heterotrimeric complexes, forming the capsid floor between the hexons and pentons, are each composed of one molecule of ORF62 and two molecules of ORF26. Each of these proteins has significant amino acid sequence homology to capsid proteins in alpha- and betaherpesviruses. In contrast, the fourth protein, ORF65, lacks significant sequence homology to its structural counterparts from the other subfamilies. Nevertheless, this small, basic, and highly antigenic protein decorates the surface of the capsids, as does, for example, the even smaller basic capsid protein VP26 of herpes simplex virus type 1. We have also found that, as with the alpha- and betaherpesviruses, lytic replication of KSHV leads to the formation of at least three capsid species, A, B, and C, with masses of approximately 200, 230, and 300 MDa, respectively. A capsids are empty, B capsids contain an inner array of a fifth structural protein, ORF17.5, and C capsids contain the viral genome.
Figures



Similar articles
-
Capsid structure of Kaposi's sarcoma-associated herpesvirus, a gammaherpesvirus, compared to those of an alphaherpesvirus, herpes simplex virus type 1, and a betaherpesvirus, cytomegalovirus.J Virol. 2001 Mar;75(6):2879-90. doi: 10.1128/JVI.75.6.2879-2890.2001. J Virol. 2001. PMID: 11222713 Free PMC article.
-
Three-dimensional localization of pORF65 in Kaposi's sarcoma-associated herpesvirus capsid.J Virol. 2003 Apr;77(7):4291-7. doi: 10.1128/jvi.77.7.4291-4297.2003. J Virol. 2003. PMID: 12634386 Free PMC article.
-
Three-dimensional structure of the human herpesvirus 8 capsid.J Virol. 2000 Oct;74(20):9646-54. doi: 10.1128/jvi.74.20.9646-9654.2000. J Virol. 2000. PMID: 11000237 Free PMC article.
-
The Terminase Complex of Each Human Herpesvirus.Biol Pharm Bull. 2024;47(5):912-916. doi: 10.1248/bpb.b23-00717. Biol Pharm Bull. 2024. PMID: 38692868 Review.
-
Assemblins as maturational proteases in herpesviruses.J Gen Virol. 2017 Aug;98(8):1969-1984. doi: 10.1099/jgv.0.000872. Epub 2017 Jul 31. J Gen Virol. 2017. PMID: 28758622 Review.
Cited by
-
The assembly domain of the small capsid protein of Kaposi's sarcoma-associated herpesvirus.J Virol. 2012 Nov;86(21):11926-30. doi: 10.1128/JVI.01430-12. Epub 2012 Aug 22. J Virol. 2012. PMID: 22915821 Free PMC article.
-
Addition of a C-terminal cysteine improves the anti-herpes simplex virus activity of a peptide containing the human immunodeficiency virus type 1 TAT protein transduction domain.Antimicrob Agents Chemother. 2007 May;51(5):1596-607. doi: 10.1128/AAC.01009-06. Epub 2007 Jan 29. Antimicrob Agents Chemother. 2007. PMID: 17261627 Free PMC article.
-
De novo infection with rhesus monkey rhadinovirus leads to the accumulation of multiple intranuclear capsid species during lytic replication but favors the release of genome-containing virions.J Virol. 2003 Dec;77(24):13439-47. doi: 10.1128/jvi.77.24.13439-13447.2003. J Virol. 2003. PMID: 14645602 Free PMC article.
-
Pervasive transcription of a herpesvirus genome generates functionally important RNAs.mBio. 2014 Mar 11;5(2):e01033-13. doi: 10.1128/mBio.01033-13. mBio. 2014. PMID: 24618256 Free PMC article.
-
Kaposi's Sarcoma-Associated Herpesvirus ORF67.5 Functions as a Component of the Terminase Complex.J Virol. 2023 Jun 29;97(6):e0047523. doi: 10.1128/jvi.00475-23. Epub 2023 Jun 5. J Virol. 2023. PMID: 37272800 Free PMC article.
References
-
- Boshoff C, Schulz T F, Kennedy M M, Graham A K, Fisher C, Thomas A, McGee J O, Weiss R A, O'Leary J J. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat Med. 1995;1:1274–1278. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

