Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees
- PMID: 11170892
- PMCID: PMC1235277
- DOI: 10.1086/318206
Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees
Abstract
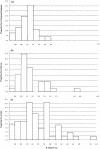
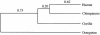
To study the genomic divergences among hominoids and to estimate the effective population size of the common ancestor of humans and chimpanzees, we selected 53 autosomal intergenic nonrepetitive DNA segments from the human genome and sequenced them in a human, a chimpanzee, a gorilla, and an orangutan. The average sequence divergence was only 1.24% +/- 0.07% for the human-chimpanzee pair, 1.62% +/- 0.08% for the human-gorilla pair, and 1.63% +/- 0.08% for the chimpanzee-gorilla pair. These estimates, which were confirmed by additional data from GenBank, are substantially lower than previous ones, which included repetitive sequences and might have been based on less-accurate sequence data. The average sequence divergences between orangutans and humans, chimpanzees, and gorillas were 3.08% +/- 0.11%, 3.12% +/- 0.11%, and 3.09% +/- 0.11%, respectively, which also are substantially lower than previous estimates. The sequence divergences in other regions between hominoids were estimated from extensive data in GenBank and the literature, and Alus showed the highest divergence, followed in order by Y-linked noncoding regions, pseudogenes, autosomal intergenic regions, X-linked noncoding regions, synonymous sites, introns, and nonsynonymous sites. The neighbor-joining tree derived from the concatenated sequence of the 53 segments--24,234 bp in length--supports the Homo-Pan clade with a 100% bootstrap value. However, when each segment is analyzed separately, 22 of the 53 segments (approximately 42%) give a tree that is incongruent with the species tree, suggesting a large effective population size (N(e)) of the common ancestor of Homo and Pan. Indeed, a parsimony analysis of the 53 segments and 37 protein-coding genes leads to an estimate of N(e) = 52,000 to 96,000. As this estimate is 5 to 9 times larger than the long-term effective population size of humans (approximately 10,000) estimated from various genetic polymorphism data, the human lineage apparently had experienced a large reduction in effective population size after its separation from the chimpanzee lineage. Our analysis assumes a molecular clock, which is in fact supported by the sequence data used. Taking the orangutan speciation date as 12 to 16 million years ago, we obtain an estimate of 4.6 to 6.2 million years for the Homo-Pan divergence and an estimate of 6.2 to 8.4 million years for the gorilla speciation date, suggesting that the gorilla lineage branched off 1.6 to 2.2 million years earlier than did the human-chimpanzee divergence.
Figures
Similar articles
-
Nucleotide sequences of immunoglobulin-epsilon pseudogenes in man and apes and their phylogenetic relationships.J Mol Biol. 1989 Jan 5;205(1):85-90. doi: 10.1016/0022-2836(89)90366-5. J Mol Biol. 1989. PMID: 2926810
-
Gorilla and orangutan c-myc nucleotide sequences: inference on hominoid phylogeny.J Mol Evol. 1995 Sep;41(3):262-76. doi: 10.1007/BF00186538. J Mol Evol. 1995. PMID: 7563112
-
Interspecific variation at the Y-linked RPS4Y locus in hominoids: implications for phylogeny.Am J Phys Anthropol. 1996 Nov;101(3):333-43. doi: 10.1002/(SICI)1096-8644(199611)101:3<333::AID-AJPA3>3.0.CO;2-#. Am J Phys Anthropol. 1996. PMID: 8922180
-
The phylogeny of the hominoid primates, as indicated by DNA-DNA hybridization.J Mol Evol. 1984;20(1):2-15. doi: 10.1007/BF02101980. J Mol Evol. 1984. PMID: 6429338 Review.
-
Reconstructing phylogenies and phenotypes: a molecular view of human evolution.J Anat. 2008 Apr;212(4):337-53. doi: 10.1111/j.1469-7580.2007.00840.x. J Anat. 2008. PMID: 18380860 Free PMC article. Review.
Cited by
-
Recent advances in understanding the role of nutrition in human genome evolution.Adv Nutr. 2011 Nov;2(6):486-96. doi: 10.3945/an.111.001024. Epub 2011 Nov 3. Adv Nutr. 2011. PMID: 22332091 Free PMC article. Review.
-
Positive selection for the male functionality of a co-retroposed gene in the hominoids.BMC Evol Biol. 2009 Oct 15;9:252. doi: 10.1186/1471-2148-9-252. BMC Evol Biol. 2009. PMID: 19832993 Free PMC article.
-
Statistical evaluation of alternative models of human evolution.Proc Natl Acad Sci U S A. 2007 Nov 6;104(45):17614-9. doi: 10.1073/pnas.0708280104. Epub 2007 Oct 31. Proc Natl Acad Sci U S A. 2007. PMID: 17978179 Free PMC article.
-
Appropriate Assignment of Fossil Calibration Information Minimizes the Difference between Phylogenetic and Pedigree Mutation Rates in Humans.Life (Basel). 2018 Oct 22;8(4):49. doi: 10.3390/life8040049. Life (Basel). 2018. PMID: 30360410 Free PMC article.
-
Human-specific duplication and mosaic transcripts: the recent paralogous structure of chromosome 22.Am J Hum Genet. 2002 Jan;70(1):83-100. doi: 10.1086/338458. Epub 2001 Nov 30. Am J Hum Genet. 2002. PMID: 11731936 Free PMC article.
References
Electronic-Database Information
-
- BLAST server, http://www.ncbi.nlm.nih.gov/BLAST/
-
- Genome Channel, http://genome.ornl.gov/GCat/species.shtml (for 53 DNA segments studied)
-
- Silver Project Home Page, http://sayer.lab.nig.ac.jp/~silver/homoNuc.html (for accession numbers of coding regions)
References
-
- Bailey WJ, Fitch DH, Tangle DA, Czelusniak J, Slightom JL, Goodman M (1991) Molecular evolution of the ψη-globin gene locus: gibbon phylogeny and the hominoid slowdown. Mol Biol Evol 8:155–184 - PubMed
-
- Bohossian HB, Skaletsky H, Page DC (2000) Unexpectedly similar rates of nucleotide substitution found in male and female hominids. Nature 406:622–625 - PubMed
-
- Dorit RL, Akashi H, Gilbert W (1995) Absence of polymorphism at the ZFY locus on the human Y chromosome. Science 268:1183–1185 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials