Self-assembly of nucleocapsid-like particles from recombinant hepatitis C virus core protein
- PMID: 11160716
- PMCID: PMC114796
- DOI: 10.1128/JVI.75.5.2119-2129.2001
Self-assembly of nucleocapsid-like particles from recombinant hepatitis C virus core protein
Abstract
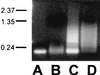
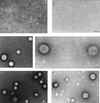
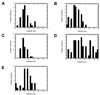
Little is known about the assembly pathway and structure of hepatitis C virus (HCV) since insufficient quantities of purified virus are available for detailed biophysical and structural studies. Here, we show that bacterially expressed HCV core proteins can efficiently self-assemble in vitro into nucleocapsid-like particles. These particles have a regular, spherical morphology with a modal distribution of diameters of approximately 60 nm. Self-assembly of nucleocapsid-like particles requires structured RNA molecules. The 124 N-terminal residues of the core protein are sufficient for self-assembly into nucleocapsid-like particles. Inclusion of the carboxy-terminal domain of the core protein modifies the core assembly pathway such that the resultant particles have an irregular outline. However, these particles are similar in size and shape to those assembled from the 124 N-terminal residues of the core protein. These results provide novel opportunities to delineate protein-protein and protein-RNA interactions critical for HCV assembly, to study the molecular details of HCV assembly, and for performing high-throughput screening of assembly inhibitors.
Figures
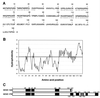
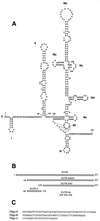
 , alpha helix.
, alpha helix.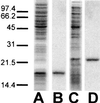

Similar articles
-
Conformational changes accompanying self-assembly of the hepatitis C virus core protein.Virology. 2002 Mar 15;294(2):239-45. doi: 10.1006/viro.2001.1325. Virology. 2002. PMID: 12009865
-
Structural requirements for assembly and homotypic interactions of the hepatitis C virus core protein.Virus Res. 2006 Dec;122(1-2):137-43. doi: 10.1016/j.virusres.2006.07.008. Epub 2006 Sep 1. Virus Res. 2006. PMID: 16949699
-
The N-terminal half of the core protein of hepatitis C virus is sufficient for nucleocapsid formation.J Gen Virol. 2004 Apr;85(Pt 4):971-981. doi: 10.1099/vir.0.79775-0. J Gen Virol. 2004. PMID: 15039539
-
[Involvement of nonstructural protein 5A and lipids on production of hepatitis C virus particles].Uirusu. 2008 Dec;58(2):199-205. doi: 10.2222/jsv.58.199. Uirusu. 2008. PMID: 19374198 Review. Japanese.
-
Assembly of hepatitis C virus particles.Microbiol Immunol. 2011 Jan;55(1):12-8. doi: 10.1111/j.1348-0421.2010.00274.x. Microbiol Immunol. 2011. PMID: 21175769 Review.
Cited by
-
Analysis of the RNA chaperoning activity of the hepatitis C virus core protein on the conserved 3'X region of the viral genome.Nucleic Acids Res. 2012 Mar;40(6):2540-53. doi: 10.1093/nar/gkr1140. Epub 2011 Nov 29. Nucleic Acids Res. 2012. PMID: 22127859 Free PMC article.
-
Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex.J Virol. 2002 Jun;76(12):5974-84. doi: 10.1128/jvi.76.12.5974-5984.2002. J Virol. 2002. PMID: 12021330 Free PMC article.
-
Activation of the Ca2+/NFAT Pathway by Assembly of Hepatitis C Virus Core Protein into Nucleocapsid-like Particles.Viruses. 2022 Apr 6;14(4):761. doi: 10.3390/v14040761. Viruses. 2022. PMID: 35458491 Free PMC article.
-
Molecular biology of hepatitis C virus.J Gastroenterol. 2007 Jun;42(6):411-23. doi: 10.1007/s00535-007-2030-3. Epub 2007 Jun 29. J Gastroenterol. 2007. PMID: 17671755 Review.
-
Targeting of hepatitis C virus core protein to mitochondria through a novel C-terminal localization motif.J Virol. 2004 Aug;78(15):7958-68. doi: 10.1128/JVI.78.15.7958-7968.2004. J Virol. 2004. PMID: 15254168 Free PMC article.
References
-
- Beard M, Abell G, Honda M, Carroll A, Gartland M, Clarke B, Susuki K, Lanford R, Sangar D V, Lemon S M. An infectious clone of a Japanese genotype 1.b. hepatitis C virus. Hepatology. 1999;30:316–324. - PubMed
-
- Bothner B, Schneemann A, Marshall D, Reddy V, Johnson J E, Siuzdak G. Crystallographically identical virus capsids display different properties in solution. Nat Struct Biol. 1999;6:114–116. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

